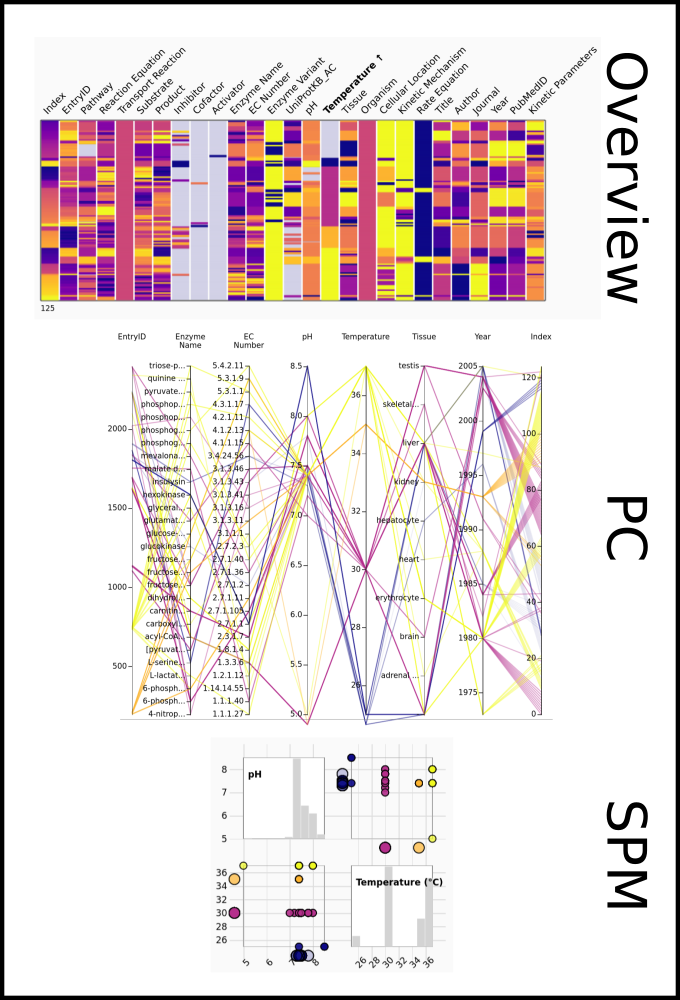
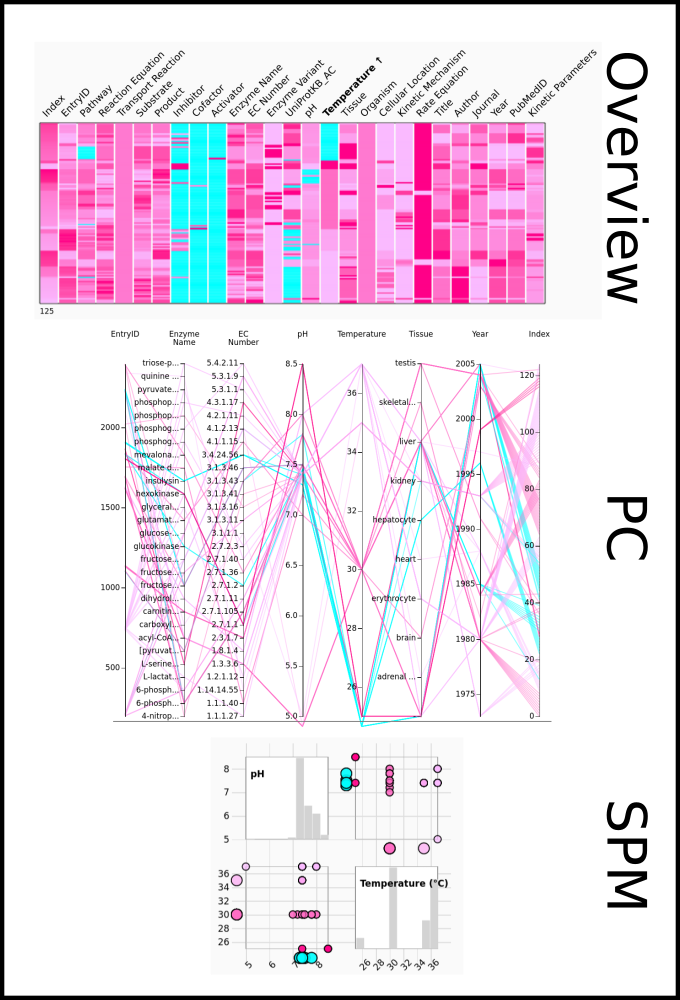
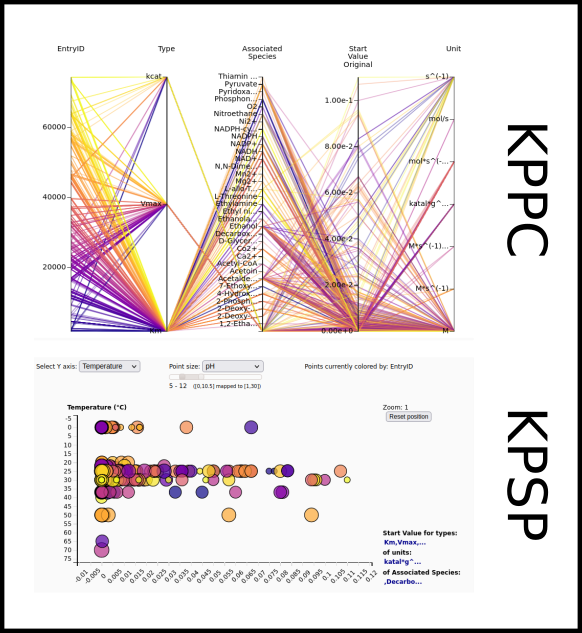
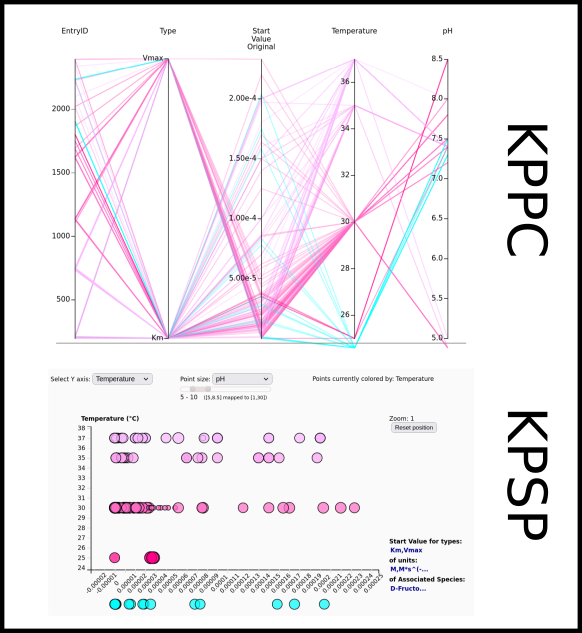
To improve SABIO-RK data interoperability, semantic markup was added to web pages as described and defined by
This structured information makes it easier to discover, collate and analyse our data.
|
64001
|
Purification, characterization, cloning, and expression of the chicken liver ecto-ATP-diphosphohydrolase
|
|
64002
|
Purification, characterization, cloning, and expression of the chicken liver ecto-ATP-diphosphohydrolase
|
|
64003
|
Purification, characterization, cloning, and expression of the chicken liver ecto-ATP-diphosphohydrolase
|
|
64004
|
Purification, characterization, cloning, and expression of the chicken liver ecto-ATP-diphosphohydrolase
|
|
64005
|
Discovery and structure determination of the orphan enzyme isoxanthopterin deaminase
|
|
64006
|
Discovery and structure determination of the orphan enzyme isoxanthopterin deaminase
|
|
64007
|
Discovery and structure determination of the orphan enzyme isoxanthopterin deaminase
|
|
64008
|
Discovery and structure determination of the orphan enzyme isoxanthopterin deaminase
|
|
64009
|
GUD1 (YDL238c) encodes Saccharomyces cerevisiae guanine deaminase, an enzyme expressed during post-diauxic ...
|
|
64010
|
Purification and characterization of rat liver GTP cyclohydrolase I. Cooperative binding of GTP to the enzyme
|
|
64011
|
Purification and characterization of rat liver GTP cyclohydrolase I. Cooperative binding of GTP to the enzyme
|
|
64012
|
Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and ...
|
|
64013
|
Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and ...
|
|
64014
|
Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and ...
|
|
64015
|
Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and ...
|
|
64016
|
Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and ...
|
|
64017
|
Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and ...
|
|
64018
|
Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and ...
|
|
64019
|
Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and ...
|
|
64020
|
Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and ...
|
|
64021
|
Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and ...
|
|
64022
|
Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and ...
|
|
64023
|
Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and ...
|
|
64024
|
Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and ...
|
|
64025
|
Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and ...
|
|
64026
|
Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and ...
|
|
64027
|
Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and ...
|
|
64028
|
Reassessment of effects on lignification and vascular development in the irx4 Arabidopsis mutant.
|
|
64029
|
Reassessment of effects on lignification and vascular development in the irx4 Arabidopsis mutant.
|
|
64030
|
Reassessment of effects on lignification and vascular development in the irx4 Arabidopsis mutant.
|
|
64031
|
Reassessment of effects on lignification and vascular development in the irx4 Arabidopsis mutant.
|
|
64032
|
Reassessment of effects on lignification and vascular development in the irx4 Arabidopsis mutant.
|
|
64033
|
Reassessment of effects on lignification and vascular development in the irx4 Arabidopsis mutant.
|
|
64034
|
Calcium ions and the regulation of NAD+-linked isocitrate dehydrogenase from the mitochondria of rat heart and ...
|
|
64035
|
Calcium ions and the regulation of NAD+-linked isocitrate dehydrogenase from the mitochondria of rat heart and ...
|
|
64036
|
Calcium ions and the regulation of NAD+-linked isocitrate dehydrogenase from the mitochondria of rat heart and ...
|
|
64037
|
Calcium ions and the regulation of NAD+-linked isocitrate dehydrogenase from the mitochondria of rat heart and ...
|
|
64038
|
Calcium ions and the regulation of NAD+-linked isocitrate dehydrogenase from the mitochondria of rat heart and ...
|
|
64039
|
Calcium ions and the regulation of NAD+-linked isocitrate dehydrogenase from the mitochondria of rat heart and ...
|
|
64040
|
Calcium ions and the regulation of NAD+-linked isocitrate dehydrogenase from the mitochondria of rat heart and ...
|
|
64041
|
Calcium ions and the regulation of NAD+-linked isocitrate dehydrogenase from the mitochondria of rat heart and ...
|
|
64042
|
Calcium ions and the regulation of NAD+-linked isocitrate dehydrogenase from the mitochondria of rat heart and ...
|
|
64043
|
Calcium ions and the regulation of NAD+-linked isocitrate dehydrogenase from the mitochondria of rat heart and ...
|
|
64044
|
Calcium ions and the regulation of NAD+-linked isocitrate dehydrogenase from the mitochondria of rat heart and ...
|
|
64045
|
Calcium ions and the regulation of NAD+-linked isocitrate dehydrogenase from the mitochondria of rat heart and ...
|
|
64046
|
Calcium ions and the regulation of NAD+-linked isocitrate dehydrogenase from the mitochondria of rat heart and ...
|
|
64047
|
Calcium ions and the regulation of NAD+-linked isocitrate dehydrogenase from the mitochondria of rat heart and ...
|
|
64048
|
The effects of adenine nucleotides on NADH binding to mitochondrial malate dehydrogenase
|
|
64049
|
NAD+/NADP+-dependent malic enzyme: evidence of a NADP+ preferring activity in human skeletal muscle.
|
|
64050
|
NAD+/NADP+-dependent malic enzyme: evidence of a NADP+ preferring activity in human skeletal muscle.
|
|
64051
|
Modulation of succinate transport in Hep G2 cell line by PKC
|
|
64052
|
Modulation of succinate transport in Hep G2 cell line by PKC
|
|
64053
|
Modulation of succinate transport in Hep G2 cell line by PKC
|
|
64054
|
Modulation of succinate transport in Hep G2 cell line by PKC
|
|
64055
|
Modulation of succinate transport in Hep G2 cell line by PKC
|
|
64056
|
Modulation of succinate transport in Hep G2 cell line by PKC
|
|
64057
|
Modulation of succinate transport in Hep G2 cell line by PKC
|
|
64058
|
Modulation of succinate transport in Hep G2 cell line by PKC
|
|
64059
|
Modulation of succinate transport in Hep G2 cell line by PKC
|
|
64060
|
Modulation of succinate transport in Hep G2 cell line by PKC
|
|
64061
|
Modulation of succinate transport in Hep G2 cell line by PKC
|
|
64062
|
Modulation of succinate transport in Hep G2 cell line by PKC
|
|
64063
|
Modulation of succinate transport in Hep G2 cell line by PKC
|
|
64064
|
Modulation of succinate transport in Hep G2 cell line by PKC
|
|
64065
|
Modulation of succinate transport in Hep G2 cell line by PKC
|
|
64066
|
Modulation of succinate transport in Hep G2 cell line by PKC
|
|
64067
|
Purification and partial structural and kinetic characterization of an aromatic L-alpha-hydroxy acid ...
|
|
64068
|
Purification and partial structural and kinetic characterization of an aromatic L-alpha-hydroxy acid ...
|
|
64069
|
Purification and partial structural and kinetic characterization of an aromatic L-alpha-hydroxy acid ...
|
|
64070
|
Purification and partial structural and kinetic characterization of an aromatic L-alpha-hydroxy acid ...
|
|
64071
|
Purification and partial structural and kinetic characterization of an aromatic L-alpha-hydroxy acid ...
|
|
64072
|
Purification and partial structural and kinetic characterization of an aromatic L-alpha-hydroxy acid ...
|
|
64073
|
Purification and partial structural and kinetic characterization of an aromatic L-alpha-hydroxy acid ...
|
|
64074
|
The NAD-linked aromatic alpha-hydroxy acid dehydrogenase from Trypanosoma cruzi. A new member of the cytosolic ...
|
|
64075
|
The NAD-linked aromatic alpha-hydroxy acid dehydrogenase from Trypanosoma cruzi. A new member of the cytosolic ...
|
|
64076
|
The NAD-linked aromatic alpha-hydroxy acid dehydrogenase from Trypanosoma cruzi. A new member of the cytosolic ...
|
|
64077
|
The NAD-linked aromatic alpha-hydroxy acid dehydrogenase from Trypanosoma cruzi. A new member of the cytosolic ...
|
|
64078
|
The NAD-linked aromatic alpha-hydroxy acid dehydrogenase from Trypanosoma cruzi. A new member of the cytosolic ...
|
|
64079
|
The NAD-linked aromatic alpha-hydroxy acid dehydrogenase from Trypanosoma cruzi. A new member of the cytosolic ...
|
|
64080
|
The NAD-linked aromatic alpha-hydroxy acid dehydrogenase from Trypanosoma cruzi. A new member of the cytosolic ...
|
|
64081
|
The NAD-linked aromatic alpha-hydroxy acid dehydrogenase from Trypanosoma cruzi. A new member of the cytosolic ...
|
|
64082
|
Molecular cloning and characterization of a malic enzyme gene from the oleaginous yeast Lipomyces starkeyi
|
|
64083
|
Molecular cloning and characterization of a malic enzyme gene from the oleaginous yeast Lipomyces starkeyi
|
|
64084
|
Molecular cloning and characterization of a malic enzyme gene from the oleaginous yeast Lipomyces starkeyi
|
|
64085
|
Molecular cloning and characterization of a malic enzyme gene from the oleaginous yeast Lipomyces starkeyi
|
|
64086
|
Crystal structure of heart 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB2) and the inhibitory ...
|
|
64087
|
Crystal structure of heart 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB2) and the inhibitory ...
|
|
64088
|
Crystal structure of heart 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB2) and the inhibitory ...
|
|
64089
|
Crystal structure of heart 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB2) and the inhibitory ...
|
|
64090
|
Identification of transport pathways for citric acid cycle intermediates in the human colon carcinoma cell ...
|
|
64091
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64092
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64093
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64094
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64095
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64096
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64097
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64098
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64099
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64100
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64101
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64102
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64103
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64104
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64105
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64106
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64107
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64108
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64109
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64110
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64111
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64112
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64113
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64114
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64115
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64116
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64117
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64118
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64119
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64120
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64121
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64122
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64123
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64124
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64125
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64126
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64127
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64128
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64129
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64130
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64131
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64132
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64133
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64134
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64135
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64136
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64137
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64138
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64139
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64140
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64141
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64142
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64143
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64144
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64145
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64146
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64147
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64148
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64149
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64150
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64151
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64152
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64153
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64154
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64155
|
A preliminary characterization of the cytosolic glutathione transferase proteome from Drosophila melanogaster.
|
|
64156
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64157
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64158
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64159
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64160
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64161
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64162
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64163
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64164
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64165
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64166
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64167
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64168
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64169
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64170
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64171
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64172
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64173
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64174
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64175
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64176
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64177
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64178
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64179
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64180
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64181
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64182
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64183
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64184
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64185
|
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium ...
|
|
64186
|
Control of citric acid cycle activity in rat heart mitochondria.
|
|
64187
|
Control of citric acid cycle activity in rat heart mitochondria.
|
|
64188
|
Control of citric acid cycle activity in rat heart mitochondria.
|
|
64189
|
Control of citric acid cycle activity in rat heart mitochondria.
|
|
64190
|
Exploring the role of L209 residue in the active site of NDM-1 a metallo-beta-lactamase
|
|
64191
|
Exploring the role of L209 residue in the active site of NDM-1 a metallo-beta-lactamase
|
|
64192
|
Exploring the role of L209 residue in the active site of NDM-1 a metallo-beta-lactamase
|
|
64193
|
Exploring the role of L209 residue in the active site of NDM-1 a metallo-beta-lactamase
|
|
64194
|
Exploring the role of L209 residue in the active site of NDM-1 a metallo-beta-lactamase
|
|
64195
|
Exploring the role of L209 residue in the active site of NDM-1 a metallo-beta-lactamase
|
|
64196
|
Exploring the role of L209 residue in the active site of NDM-1 a metallo-beta-lactamase
|
|
64197
|
Purification and properties of the NAD(P)-dependent malic enzyme from human placental mitochondria
|
|
64198
|
Purification and properties of the NAD(P)-dependent malic enzyme from human placental mitochondria
|
|
64199
|
Purification and properties of the NAD(P)-dependent malic enzyme from human placental mitochondria
|
|
64200
|
Purification and properties of the NAD(P)-dependent malic enzyme from human placental mitochondria
|
|
64201
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64202
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64203
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64204
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64205
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64206
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64207
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64208
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64209
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64210
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64211
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64212
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64213
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64214
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64215
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64216
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64217
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64218
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64219
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64220
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64221
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64222
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64223
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64224
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64225
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64226
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64227
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64228
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64229
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64230
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64231
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64232
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64233
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64234
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64235
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64236
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64237
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64238
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64239
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64240
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64241
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64242
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64243
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64244
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64245
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64246
|
Molecular mechanism of the allosteric regulation of the αγ heterodimer of human NAD-dependent ...
|
|
64247
|
Isotope and solvent effects of deuterium on aconitase.
|
|
64248
|
Isotope and solvent effects of deuterium on aconitase.
|
|
64249
|
Isotope and solvent effects of deuterium on aconitase.
|
|
64250
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64251
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64252
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64253
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64254
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64255
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64256
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64257
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64258
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64259
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64260
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64261
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64262
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64263
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64264
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64265
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64266
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64267
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64268
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64269
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64270
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64271
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64272
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64273
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64274
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64275
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64276
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64277
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64278
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64279
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64280
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64281
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64282
|
Rationale for reclassification of a distinctive subdivision of mammalian class Mu glutathione S-transferases ...
|
|
64283
|
Structure and mechanism of spermidine synthases
|
|
64284
|
Structure and mechanism of spermidine synthases
|
|
64285
|
Structure and mechanism of spermidine synthases
|
|
64286
|
Structure and mechanism of spermidine synthases
|
|
64287
|
Structure and mechanism of spermidine synthases
|
|
64288
|
Structure and mechanism of spermidine synthases
|
|
64289
|
Structure and mechanism of spermidine synthases
|
|
64290
|
Structure and mechanism of spermidine synthases
|
|
64291
|
Structure and mechanism of spermidine synthases
|
|
64292
|
Structure and mechanism of spermidine synthases
|
|
64293
|
Structure and mechanism of spermidine synthases
|
|
64294
|
Engineering broad-spectrum digestion of polyuronides from an exolytic polysaccharide lyase
|
|
64295
|
Engineering broad-spectrum digestion of polyuronides from an exolytic polysaccharide lyase
|
|
64296
|
Engineering broad-spectrum digestion of polyuronides from an exolytic polysaccharide lyase
|
|
64297
|
Engineering broad-spectrum digestion of polyuronides from an exolytic polysaccharide lyase
|
|
64298
|
Engineering broad-spectrum digestion of polyuronides from an exolytic polysaccharide lyase
|
|
64299
|
Engineering broad-spectrum digestion of polyuronides from an exolytic polysaccharide lyase
|
|
64300
|
Engineering broad-spectrum digestion of polyuronides from an exolytic polysaccharide lyase
|
|
64301
|
Engineering broad-spectrum digestion of polyuronides from an exolytic polysaccharide lyase
|
|
64302
|
Engineering broad-spectrum digestion of polyuronides from an exolytic polysaccharide lyase
|
|
64303
|
Engineering broad-spectrum digestion of polyuronides from an exolytic polysaccharide lyase
|
|
64304
|
Synthesis and biological evaluation of methanesulfonamide analogues of rofecoxib: Replacement of ...
|
|
64305
|
Synthesis and biological evaluation of methanesulfonamide analogues of rofecoxib: Replacement of ...
|
|
64306
|
Synthesis and biological evaluation of methanesulfonamide analogues of rofecoxib: Replacement of ...
|
|
64307
|
Synthesis and biological evaluation of methanesulfonamide analogues of rofecoxib: Replacement of ...
|
|
64308
|
Synthesis and biological evaluation of methanesulfonamide analogues of rofecoxib: Replacement of ...
|
|
64309
|
Synthesis and biological evaluation of methanesulfonamide analogues of rofecoxib: Replacement of ...
|
|
64310
|
Synthesis and biological evaluation of methanesulfonamide analogues of rofecoxib: Replacement of ...
|
|
64311
|
Synthesis and biological evaluation of methanesulfonamide analogues of rofecoxib: Replacement of ...
|
|
64312
|
Synthesis and biological evaluation of methanesulfonamide analogues of rofecoxib: Replacement of ...
|
|
64313
|
Synthesis and biological evaluation of methanesulfonamide analogues of rofecoxib: Replacement of ...
|
|
64314
|
Synthesis and biological evaluation of methanesulfonamide analogues of rofecoxib: Replacement of ...
|
|
64315
|
Conformational changes associated with cofactor/substrate binding of 6-phosphogluconate dehydrogenase from ...
|
|
64316
|
Conformational changes associated with cofactor/substrate binding of 6-phosphogluconate dehydrogenase from ...
|
|
64317
|
Conformational changes associated with cofactor/substrate binding of 6-phosphogluconate dehydrogenase from ...
|
|
64318
|
Conformational changes associated with cofactor/substrate binding of 6-phosphogluconate dehydrogenase from ...
|
|
64319
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64320
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64321
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64322
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64323
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64324
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64325
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64326
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64327
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64328
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64329
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64330
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64331
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64332
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64333
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64334
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64335
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64336
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64337
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64338
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64339
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64340
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64341
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64342
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64343
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64344
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64345
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64346
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64347
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64348
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64349
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64350
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64351
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64352
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64353
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64354
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64355
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64356
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64357
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64358
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64359
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64360
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64361
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64362
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64363
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64364
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64365
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64366
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64367
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64368
|
SUCCINIC THIOKINASE. II. KINETIC STUDIES: INITIAL VELOCITY, PRODUCT INHIBITION, AND EFFECT OF ARSENATE
|
|
64369
|
X-ray crystallographic and kinetic correlation of a clinically observed human fumarase mutation
|
|
64370
|
X-ray crystallographic and kinetic correlation of a clinically observed human fumarase mutation
|
|
64371
|
X-ray crystallographic and kinetic correlation of a clinically observed human fumarase mutation
|
|
64372
|
X-ray crystallographic and kinetic correlation of a clinically observed human fumarase mutation
|
|
64373
|
Triphosphopyridine nucleotide-linked aldehyde dehydrogenase from yeast.
|
|
64374
|
Triphosphopyridine nucleotide-linked aldehyde dehydrogenase from yeast.
|
|
64375
|
Triphosphopyridine nucleotide-linked aldehyde dehydrogenase from yeast.
|
|
64376
|
Human 2-oxoglutarate dehydrogenase complex E1 component forms a thiamin-derived radical by aerobic oxidation ...
|
|
64377
|
Human 2-oxoglutarate dehydrogenase complex E1 component forms a thiamin-derived radical by aerobic oxidation ...
|
|
64378
|
The three-dimensional structural basis of type II hyperprolinemia.
|
|
64379
|
The three-dimensional structural basis of type II hyperprolinemia.
|
|
64380
|
The three-dimensional structural basis of type II hyperprolinemia.
|
|
64381
|
The three-dimensional structural basis of type II hyperprolinemia.
|
|
64382
|
Effects of aluminum on activity of krebs cycle enzymes and glutamate dehydrogenase in rat brain homogenate.
|
|
64383
|
Effects of aluminum on activity of krebs cycle enzymes and glutamate dehydrogenase in rat brain homogenate.
|
|
64384
|
Effects of aluminum on activity of krebs cycle enzymes and glutamate dehydrogenase in rat brain homogenate.
|
|
64385
|
Effects of aluminum on activity of krebs cycle enzymes and glutamate dehydrogenase in rat brain homogenate.
|
|
64386
|
Effects of aluminum on activity of krebs cycle enzymes and glutamate dehydrogenase in rat brain homogenate.
|
|
64387
|
Effects of aluminum on activity of krebs cycle enzymes and glutamate dehydrogenase in rat brain homogenate.
|
|
64388
|
Effects of aluminum on activity of krebs cycle enzymes and glutamate dehydrogenase in rat brain homogenate.
|
|
64389
|
Effects of aluminum on activity of krebs cycle enzymes and glutamate dehydrogenase in rat brain homogenate.
|
|
64390
|
Yeast aldehyde dehydrogenase. II. Properties of the homogeneous enzyme preparations.
|
|
64391
|
Yeast aldehyde dehydrogenase. II. Properties of the homogeneous enzyme preparations.
|
|
64392
|
Yeast aldehyde dehydrogenase. II. Properties of the homogeneous enzyme preparations.
|
|
64393
|
Yeast aldehyde dehydrogenase. II. Properties of the homogeneous enzyme preparations.
|
|
64394
|
Yeast aldehyde dehydrogenase. II. Properties of the homogeneous enzyme preparations.
|
|
64395
|
Yeast aldehyde dehydrogenase. II. Properties of the homogeneous enzyme preparations.
|
|
64396
|
Yeast aldehyde dehydrogenase. II. Properties of the homogeneous enzyme preparations.
|
|
64397
|
Yeast aldehyde dehydrogenase. II. Properties of the homogeneous enzyme preparations.
|
|
64398
|
Yeast aldehyde dehydrogenase. II. Properties of the homogeneous enzyme preparations.
|
|
64399
|
Yeast aldehyde dehydrogenase. II. Properties of the homogeneous enzyme preparations.
|
|
64400
|
Yeast aldehyde dehydrogenase. II. Properties of the homogeneous enzyme preparations.
|
|
64401
|
Yeast aldehyde dehydrogenase. II. Properties of the homogeneous enzyme preparations.
|
|
64402
|
Yeast aldehyde dehydrogenase. II. Properties of the homogeneous enzyme preparations.
|
|
64403
|
Yeast aldehyde dehydrogenase. II. Properties of the homogeneous enzyme preparations.
|
|
64404
|
Yeast aldehyde dehydrogenase. II. Properties of the homogeneous enzyme preparations.
|
|
64405
|
Yeast aldehyde dehydrogenase. II. Properties of the homogeneous enzyme preparations.
|
|
64406
|
Yeast aldehyde dehydrogenase. II. Properties of the homogeneous enzyme preparations.
|
|
64407
|
Yeast aldehyde dehydrogenase. II. Properties of the homogeneous enzyme preparations.
|
|
64408
|
Yeast aldehyde dehydrogenase. II. Properties of the homogeneous enzyme preparations.
|
|
64409
|
The purification and some properties of the Mg(2+)-activated cytosolic aldehyde dehydrogenase of Saccharomyces ...
|
|
64410
|
The purification and some properties of the Mg(2+)-activated cytosolic aldehyde dehydrogenase of Saccharomyces ...
|
|
64411
|
The purification and some properties of the Mg(2+)-activated cytosolic aldehyde dehydrogenase of Saccharomyces ...
|
|
64412
|
The purification and some properties of the Mg(2+)-activated cytosolic aldehyde dehydrogenase of Saccharomyces ...
|
|
64413
|
Expression of cDNA sequences encoding mature and precursor forms of human dihydrolipoamide dehydrogenase in ...
|
|
64414
|
Expression of cDNA sequences encoding mature and precursor forms of human dihydrolipoamide dehydrogenase in ...
|
|
64415
|
Expression of cDNA sequences encoding mature and precursor forms of human dihydrolipoamide dehydrogenase in ...
|
|
64416
|
Expression of cDNA sequences encoding mature and precursor forms of human dihydrolipoamide dehydrogenase in ...
|
|
64417
|
Increase in the stoichiometry of the functioning active sites of horse liver aldehyde dehydrogenase in the ...
|
|
64418
|
Increase in the stoichiometry of the functioning active sites of horse liver aldehyde dehydrogenase in the ...
|
|
64419
|
Increase in the stoichiometry of the functioning active sites of horse liver aldehyde dehydrogenase in the ...
|
|
64420
|
Increase in the stoichiometry of the functioning active sites of horse liver aldehyde dehydrogenase in the ...
|
|
64421
|
Increase in the stoichiometry of the functioning active sites of horse liver aldehyde dehydrogenase in the ...
|
|
64422
|
Increase in the stoichiometry of the functioning active sites of horse liver aldehyde dehydrogenase in the ...
|
|
64423
|
Nicotinamide adenine dinucleotide activation of the esterase reaction of horse liver aldehyde dehydrogenase.
|
|
64424
|
Nicotinamide adenine dinucleotide activation of the esterase reaction of horse liver aldehyde dehydrogenase.
|
|
64425
|
Nicotinamide adenine dinucleotide activation of the esterase reaction of horse liver aldehyde dehydrogenase.
|
|
64426
|
The inhibitory effects of lipoic compounds on mammalian pyruvate dehydrogenase complex and its catalytic ...
|
|
64427
|
The inhibitory effects of lipoic compounds on mammalian pyruvate dehydrogenase complex and its catalytic ...
|
|
64428
|
The inhibitory effects of lipoic compounds on mammalian pyruvate dehydrogenase complex and its catalytic ...
|
|
64429
|
The inhibitory effects of lipoic compounds on mammalian pyruvate dehydrogenase complex and its catalytic ...
|
|
64430
|
Characterization of homoisocitrate dehydrogenase involved in lysine biosynthesis of an extremely thermophilic ...
|
|
64431
|
Characterization of homoisocitrate dehydrogenase involved in lysine biosynthesis of an extremely thermophilic ...
|
|
64432
|
Characterization of homoisocitrate dehydrogenase involved in lysine biosynthesis of an extremely thermophilic ...
|
|
64433
|
Characterization of homoisocitrate dehydrogenase involved in lysine biosynthesis of an extremely thermophilic ...
|
|
64434
|
Isolation of a new dual-functional caffeine synthase gene encoding an enzyme for the conversion of ...
|
|
64435
|
Isolation of a new dual-functional caffeine synthase gene encoding an enzyme for the conversion of ...
|
|
64436
|
Isolation of a new dual-functional caffeine synthase gene encoding an enzyme for the conversion of ...
|
|
64437
|
Isolation of a new dual-functional caffeine synthase gene encoding an enzyme for the conversion of ...
|
|
64438
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64439
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64440
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64441
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64442
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64443
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64444
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64445
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64446
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64447
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64448
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64449
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64450
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64451
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64452
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64453
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64454
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64455
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64456
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64457
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64458
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64459
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64460
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64461
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64462
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64463
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64464
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64465
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64466
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64467
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64468
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64469
|
A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: ...
|
|
64470
|
Purification and characterization of malate dehydrogenase from Cryptococcus neoformans.
|
|
64471
|
Purification and characterization of malate dehydrogenase from Cryptococcus neoformans.
|
|
64472
|
Purification and characterization of malate dehydrogenase from Cryptococcus neoformans.
|
|
64473
|
Purification and characterization of malate dehydrogenase from Cryptococcus neoformans.
|
|
64474
|
Purification and characterization of malate dehydrogenase from Cryptococcus neoformans.
|
|
64475
|
Purification and characterization of malate dehydrogenase from Cryptococcus neoformans.
|
|
64476
|
Purification and characterization of malate dehydrogenase from Cryptococcus neoformans.
|
|
64477
|
Purification and characterization of malate dehydrogenase from Cryptococcus neoformans.
|
|
64478
|
Purification and characterization of malate dehydrogenase from Cryptococcus neoformans.
|
|
64479
|
Purification and characterization of malate dehydrogenase from Cryptococcus neoformans.
|
|
64480
|
Purification and characterization of malate dehydrogenase from Cryptococcus neoformans.
|
|
64481
|
Purification and characterization of malate dehydrogenase from Cryptococcus neoformans.
|
|
64482
|
Purification and characterization of malate dehydrogenase from Cryptococcus neoformans.
|
|
64483
|
Purification and characterization of malate dehydrogenase from Cryptococcus neoformans.
|
|
64484
|
Purification and characterization of malate dehydrogenase from Cryptococcus neoformans.
|
|
64485
|
A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium ...
|
|
64486
|
A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium ...
|
|
64487
|
A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium ...
|
|
64488
|
A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium ...
|
|
64489
|
Molecular characterisation of a Rhodococcus ohp operon.
|
|
64490
|
Molecular characterisation of a Rhodococcus ohp operon.
|
|
64491
|
Molecular characterisation of a Rhodococcus ohp operon.
|
|
64492
|
Chlorite dismutase from Ideonella dechloratans.
|
|
64493
|
Expression of rat liver tryptophan 2,3-dioxygenase in Escherichia coli: structural and functional ...
|
|
64494
|
Evidence for two distinct mitochondrial malic enzymes in human skeletal muscle: purification and properties of ...
|
|
64495
|
Evidence for two distinct mitochondrial malic enzymes in human skeletal muscle: purification and properties of ...
|
|
64496
|
Evidence for two distinct mitochondrial malic enzymes in human skeletal muscle: purification and properties of ...
|
|
64497
|
Evidence for two distinct mitochondrial malic enzymes in human skeletal muscle: purification and properties of ...
|
|
64498
|
Evidence for two distinct mitochondrial malic enzymes in human skeletal muscle: purification and properties of ...
|
|
64499
|
Evidence for two distinct mitochondrial malic enzymes in human skeletal muscle: purification and properties of ...
|
|
64500
|
Evidence for two distinct mitochondrial malic enzymes in human skeletal muscle: purification and properties of ...
|
|
64501
|
Evidence for two distinct mitochondrial malic enzymes in human skeletal muscle: purification and properties of ...
|
|
64502
|
Evidence for two distinct mitochondrial malic enzymes in human skeletal muscle: purification and properties of ...
|
|
64503
|
Evidence for two distinct mitochondrial malic enzymes in human skeletal muscle: purification and properties of ...
|
|
64504
|
Purification and properties of cytosolic malic enzyme from human skeletal muscle.
|
|
64505
|
Purification and properties of cytosolic malic enzyme from human skeletal muscle.
|
|
64506
|
Purification and properties of cytosolic malic enzyme from human skeletal muscle.
|
|
64507
|
Purification and properties of cytosolic malic enzyme from human skeletal muscle.
|
|
64508
|
Purification and properties of cytosolic malic enzyme from human skeletal muscle.
|
|
64509
|
Purification and properties of cytosolic malic enzyme from human skeletal muscle.
|
|
64510
|
Purification and properties of cytosolic malic enzyme from human skeletal muscle.
|
|
64511
|
Purification and properties of cytosolic malic enzyme from human skeletal muscle.
|
|
64512
|
Population and biochemical genetics of the human mitochondrial malic enzyme.
|
|
64513
|
Population and biochemical genetics of the human mitochondrial malic enzyme.
|
|
64514
|
Population and biochemical genetics of the human mitochondrial malic enzyme.
|
|
64515
|
Population and biochemical genetics of the human mitochondrial malic enzyme.
|
|
64516
|
Population and biochemical genetics of the human mitochondrial malic enzyme.
|
|
64517
|
Population and biochemical genetics of the human mitochondrial malic enzyme.
|
|
64518
|
Purification and characterization of the cytosolic NADP(+)-dependent malic enzyme from human breast cancer ...
|
|
64519
|
Purification and characterization of the cytosolic NADP(+)-dependent malic enzyme from human breast cancer ...
|
|
64520
|
Characterization and evolution of vertebrate indoleamine 2, 3-dioxygenases IDOs from monotremes and ...
|
|
64521
|
Characterization and evolution of vertebrate indoleamine 2, 3-dioxygenases IDOs from monotremes and ...
|
|
64522
|
Characterization and evolution of vertebrate indoleamine 2, 3-dioxygenases IDOs from monotremes and ...
|
|
64523
|
Characterization and evolution of vertebrate indoleamine 2, 3-dioxygenases IDOs from monotremes and ...
|
|
64524
|
Characterization and evolution of vertebrate indoleamine 2, 3-dioxygenases IDOs from monotremes and ...
|
|
64525
|
Characterization and evolution of vertebrate indoleamine 2, 3-dioxygenases IDOs from monotremes and ...
|
|
64526
|
Characterization and evolution of vertebrate indoleamine 2, 3-dioxygenases IDOs from monotremes and ...
|
|
64527
|
1-L-methyltryptophan is a more effective inhibitor of vertebrate IDO2 enzymes than 1-D-methyltryptophan.
|
|
64528
|
1-L-methyltryptophan is a more effective inhibitor of vertebrate IDO2 enzymes than 1-D-methyltryptophan.
|
|
64529
|
1-L-methyltryptophan is a more effective inhibitor of vertebrate IDO2 enzymes than 1-D-methyltryptophan.
|
|
64530
|
1-L-methyltryptophan is a more effective inhibitor of vertebrate IDO2 enzymes than 1-D-methyltryptophan.
|
|
64531
|
1-L-methyltryptophan is a more effective inhibitor of vertebrate IDO2 enzymes than 1-D-methyltryptophan.
|
|
64532
|
1-L-methyltryptophan is a more effective inhibitor of vertebrate IDO2 enzymes than 1-D-methyltryptophan.
|
|
64533
|
1-L-methyltryptophan is a more effective inhibitor of vertebrate IDO2 enzymes than 1-D-methyltryptophan.
|
|
64534
|
1-L-methyltryptophan is a more effective inhibitor of vertebrate IDO2 enzymes than 1-D-methyltryptophan.
|
|
64535
|
1-L-methyltryptophan is a more effective inhibitor of vertebrate IDO2 enzymes than 1-D-methyltryptophan.
|
|
64536
|
1-L-methyltryptophan is a more effective inhibitor of vertebrate IDO2 enzymes than 1-D-methyltryptophan.
|
|
64537
|
1-L-methyltryptophan is a more effective inhibitor of vertebrate IDO2 enzymes than 1-D-methyltryptophan.
|
|
64538
|
1-L-methyltryptophan is a more effective inhibitor of vertebrate IDO2 enzymes than 1-D-methyltryptophan.
|
|
64539
|
1-L-methyltryptophan is a more effective inhibitor of vertebrate IDO2 enzymes than 1-D-methyltryptophan.
|
|
64540
|
1-L-methyltryptophan is a more effective inhibitor of vertebrate IDO2 enzymes than 1-D-methyltryptophan.
|
|
64541
|
1-L-methyltryptophan is a more effective inhibitor of vertebrate IDO2 enzymes than 1-D-methyltryptophan.
|
|
64542
|
1-L-methyltryptophan is a more effective inhibitor of vertebrate IDO2 enzymes than 1-D-methyltryptophan.
|
|
64543
|
1-L-methyltryptophan is a more effective inhibitor of vertebrate IDO2 enzymes than 1-D-methyltryptophan.
|
|
64544
|
Mouse and human indoleamine 2,3-dioxygenase display some distinct biochemical and structural properties.
|
|
64545
|
Mouse and human indoleamine 2,3-dioxygenase display some distinct biochemical and structural properties.
|
|
64546
|
Mouse and human indoleamine 2,3-dioxygenase display some distinct biochemical and structural properties.
|
|
64547
|
Mouse and human indoleamine 2,3-dioxygenase display some distinct biochemical and structural properties.
|
|
64548
|
Mouse and human indoleamine 2,3-dioxygenase display some distinct biochemical and structural properties.
|
|
64549
|
Mouse and human indoleamine 2,3-dioxygenase display some distinct biochemical and structural properties.
|
|
64550
|
Mouse and human indoleamine 2,3-dioxygenase display some distinct biochemical and structural properties.
|
|
64551
|
Mouse and human indoleamine 2,3-dioxygenase display some distinct biochemical and structural properties.
|
|
64552
|
Mouse and human indoleamine 2,3-dioxygenase display some distinct biochemical and structural properties.
|
|
64553
|
Mouse and human indoleamine 2,3-dioxygenase display some distinct biochemical and structural properties.
|
|
64554
|
Comparative analysis of the reduction of oxaloacetate by human hepatoma and normal liver extracts.
|
|
64555
|
Comparative analysis of the reduction of oxaloacetate by human hepatoma and normal liver extracts.
|
|
64556
|
Duck liver malic enzyme: sequence of a tryptic peptide containing the cysteine residue labeled by the ...
|
|
64557
|
Duck liver malic enzyme: sequence of a tryptic peptide containing the cysteine residue labeled by the ...
|
|
64558
|
Duck liver malic enzyme: sequence of a tryptic peptide containing the cysteine residue labeled by the ...
|
|
64559
|
Duck liver malic enzyme: sequence of a tryptic peptide containing the cysteine residue labeled by the ...
|
|
64560
|
Duck liver malic enzyme: sequence of a tryptic peptide containing the cysteine residue labeled by the ...
|
|
64561
|
Purification and characterization of human liver branched-chain alpha-keto acid dehydrogenase complex.
|
|
64562
|
Purification and characterization of human liver branched-chain alpha-keto acid dehydrogenase complex.
|
|
64563
|
Purification and characterization of human liver branched-chain alpha-keto acid dehydrogenase complex.
|
|
64564
|
Purification and characterization of human liver branched-chain alpha-keto acid dehydrogenase complex.
|
|
64565
|
Purification and characterization of human liver branched-chain alpha-keto acid dehydrogenase complex.
|
|
64566
|
Kinetic characterization of the pyruvate and oxoglutarate dehydrogenase complexes from human heart.
|
|
64567
|
Kinetic characterization of the pyruvate and oxoglutarate dehydrogenase complexes from human heart.
|
|
64568
|
Kinetic characterization of the pyruvate and oxoglutarate dehydrogenase complexes from human heart.
|
|
64569
|
Kinetic characterization of the pyruvate and oxoglutarate dehydrogenase complexes from human heart.
|
|
64570
|
Kinetic characterization of the pyruvate and oxoglutarate dehydrogenase complexes from human heart.
|
|
64571
|
Kinetic characterization of the pyruvate and oxoglutarate dehydrogenase complexes from human heart.
|
|
64572
|
Kinetic characterization of the pyruvate and oxoglutarate dehydrogenase complexes from human heart.
|
|
64573
|
Kinetic characterization of the pyruvate and oxoglutarate dehydrogenase complexes from human heart.
|
|
64574
|
Kinetic characterization of the pyruvate and oxoglutarate dehydrogenase complexes from human heart.
|
|
64575
|
Kinetic characterization of the pyruvate and oxoglutarate dehydrogenase complexes from human heart.
|
|
64576
|
Kinetic characterization of the pyruvate and oxoglutarate dehydrogenase complexes from human heart.
|
|
64577
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64578
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64579
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64580
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64581
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64582
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64583
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64584
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64585
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64586
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64587
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64588
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64589
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64590
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64591
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64592
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64593
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64594
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64595
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64596
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64597
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64598
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64599
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64600
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64601
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64602
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64603
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64604
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64605
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64606
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64607
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64608
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64609
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64610
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64611
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64612
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64613
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64614
|
Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling ...
|
|
64615
|
Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase
|
|
64616
|
Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase
|
|
64617
|
Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase
|
|
64618
|
Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase
|
|
64619
|
Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase
|
|
64620
|
Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase
|
|
64621
|
Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase
|
|
64622
|
Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase
|
|
64623
|
Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase
|
|
64624
|
Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase
|
|
64625
|
Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase
|
|
64626
|
Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase
|
|
64627
|
Identification in the mould Hypocrea jecorina of a gene encoding an NADP(+): d-xylose dehydrogenase
|
|
64628
|
Identification in the mould Hypocrea jecorina of a gene encoding an NADP(+): d-xylose dehydrogenase
|
|
64629
|
Identification in the mould Hypocrea jecorina of a gene encoding an NADP(+): d-xylose dehydrogenase
|
|
64630
|
Identification in the mould Hypocrea jecorina of a gene encoding an NADP(+): d-xylose dehydrogenase
|
|
64631
|
Identification in the mould Hypocrea jecorina of a gene encoding an NADP(+): d-xylose dehydrogenase
|
|
64632
|
Identification in the mould Hypocrea jecorina of a gene encoding an NADP(+): d-xylose dehydrogenase
|
|
64633
|
Identification in the mould Hypocrea jecorina of a gene encoding an NADP(+): d-xylose dehydrogenase
|
|
64634
|
DNA topoisomerase II from Drosophila melanogaster. Relaxation of supercoiled DNA
|
|
64635
|
DNA topoisomerase II from Drosophila melanogaster. Relaxation of supercoiled DNA
|
|
64636
|
DNA topoisomerase II from Drosophila melanogaster. Relaxation of supercoiled DNA
|
|
64637
|
DNA topoisomerase II from Drosophila melanogaster. Relaxation of supercoiled DNA
|
|
64638
|
DNA topoisomerase II from Drosophila melanogaster. Relaxation of supercoiled DNA
|
|
64639
|
DNA topoisomerase II from Drosophila melanogaster. Relaxation of supercoiled DNA
|
|
64640
|
DNA topoisomerase II from Drosophila melanogaster. Relaxation of supercoiled DNA
|
|
64641
|
DNA topoisomerase II from Drosophila melanogaster. Relaxation of supercoiled DNA
|
|
64642
|
DNA topoisomerase II from Drosophila melanogaster. Relaxation of supercoiled DNA
|
|
64643
|
DNA topoisomerase II from Drosophila melanogaster. Relaxation of supercoiled DNA
|
|
64644
|
DNA topoisomerase II from Drosophila melanogaster. Relaxation of supercoiled DNA
|
|
64645
|
DNA topoisomerase II from Drosophila melanogaster. Relaxation of supercoiled DNA
|
|
64646
|
DNA topoisomerase II from Drosophila melanogaster. Relaxation of supercoiled DNA
|
|
64647
|
DNA topoisomerase II from Drosophila melanogaster. Relaxation of supercoiled DNA
|
|
64648
|
DNA topoisomerase II from Drosophila melanogaster. Relaxation of supercoiled DNA
|
|
64649
|
DNA topoisomerase II from Drosophila melanogaster. Relaxation of supercoiled DNA
|
|
64650
|
DNA topoisomerase II from Drosophila melanogaster. Relaxation of supercoiled DNA
|
|
64651
|
DNA topoisomerase II from Drosophila melanogaster. Relaxation of supercoiled DNA
|
|
64652
|
DNA topoisomerase II from Drosophila melanogaster. Relaxation of supercoiled DNA
|
|
64653
|
DNA topoisomerase II from Drosophila melanogaster. Relaxation of supercoiled DNA
|
|
64654
|
DNA topoisomerase II from Drosophila melanogaster. Relaxation of supercoiled DNA
|
|
64655
|
DNA topoisomerase II from Drosophila melanogaster. Relaxation of supercoiled DNA
|
|
64656
|
DNA topoisomerase II from Drosophila melanogaster. Relaxation of supercoiled DNA
|
|
64657
|
Single-molecule analysis of DNA uncoiling by a type II topoisomerase.
|
|
64658
|
Variation on a theme of SDR. dTDP-6-deoxy-L- lyxo-4-hexulose reductase (RmlD) shows a new Mg2+-dependent ...
|
|
64659
|
Variation on a theme of SDR. dTDP-6-deoxy-L- lyxo-4-hexulose reductase (RmlD) shows a new Mg2+-dependent ...
|
|
64660
|
Variation on a theme of SDR. dTDP-6-deoxy-L- lyxo-4-hexulose reductase (RmlD) shows a new Mg2+-dependent ...
|
|
64661
|
Structure and function of the dihydropteroate synthase from Staphylococcus aureus.
|
|
64662
|
Characterization of a (2R,3R)-2,3-butanediol dehydrogenase as the Saccharomyces cerevisiae YAL060W gene ...
|
|
64663
|
Characterization of a (2R,3R)-2,3-butanediol dehydrogenase as the Saccharomyces cerevisiae YAL060W gene ...
|
|
64664
|
Characterization of a (2R,3R)-2,3-butanediol dehydrogenase as the Saccharomyces cerevisiae YAL060W gene ...
|
|
64665
|
Characterization of a (2R,3R)-2,3-butanediol dehydrogenase as the Saccharomyces cerevisiae YAL060W gene ...
|
|
64666
|
Characterization of a (2R,3R)-2,3-butanediol dehydrogenase as the Saccharomyces cerevisiae YAL060W gene ...
|
|
64667
|
Characterization of a (2R,3R)-2,3-butanediol dehydrogenase as the Saccharomyces cerevisiae YAL060W gene ...
|
|
64668
|
Design, Synthesis and Biological Evaluation of New 1, 4-Dihydropyridine (DHP) Derivatives as Selective ...
|
|
64669
|
Design, Synthesis and Biological Evaluation of New 1, 4-Dihydropyridine (DHP) Derivatives as Selective ...
|
|
64670
|
Design, Synthesis and Biological Evaluation of New 1, 4-Dihydropyridine (DHP) Derivatives as Selective ...
|
|
64671
|
Design, Synthesis and Biological Evaluation of New 1, 4-Dihydropyridine (DHP) Derivatives as Selective ...
|
|
64672
|
Design, Synthesis and Biological Evaluation of New 1, 4-Dihydropyridine (DHP) Derivatives as Selective ...
|
|
64673
|
Design, Synthesis and Biological Evaluation of New 1, 4-Dihydropyridine (DHP) Derivatives as Selective ...
|
|
64674
|
Design, Synthesis and Biological Evaluation of New 1, 4-Dihydropyridine (DHP) Derivatives as Selective ...
|
|
64675
|
Design, Synthesis and Biological Evaluation of New 1, 4-Dihydropyridine (DHP) Derivatives as Selective ...
|
|
64676
|
Design, Synthesis and Biological Evaluation of New 1, 4-Dihydropyridine (DHP) Derivatives as Selective ...
|
|
64677
|
Design, Synthesis and Biological Evaluation of New 1, 4-Dihydropyridine (DHP) Derivatives as Selective ...
|
|
64678
|
Design, Synthesis and Biological Evaluation of New 1, 4-Dihydropyridine (DHP) Derivatives as Selective ...
|
|
64679
|
Design, Synthesis and Biological Evaluation of New 1, 4-Dihydropyridine (DHP) Derivatives as Selective ...
|
|
64680
|
Design, Synthesis and Biological Evaluation of New 1, 4-Dihydropyridine (DHP) Derivatives as Selective ...
|
|
64681
|
Design, Synthesis and Biological Evaluation of New 1, 4-Dihydropyridine (DHP) Derivatives as Selective ...
|
|
64682
|
Design, Synthesis and Biological Evaluation of New 1, 4-Dihydropyridine (DHP) Derivatives as Selective ...
|
|
64683
|
Design, Synthesis and Biological Evaluation of New 1, 4-Dihydropyridine (DHP) Derivatives as Selective ...
|
|
64684
|
Design, Synthesis and Biological Evaluation of New 1, 4-Dihydropyridine (DHP) Derivatives as Selective ...
|
|
64685
|
Design, Synthesis and Biological Evaluation of New 1, 4-Dihydropyridine (DHP) Derivatives as Selective ...
|
|
64686
|
3-Isopropylmalate is the major endogenous substrate of the Saccharomyces cerevisiae trans-aconitate ...
|
|
64687
|
3-Isopropylmalate is the major endogenous substrate of the Saccharomyces cerevisiae trans-aconitate ...
|
|
64688
|
3-Isopropylmalate is the major endogenous substrate of the Saccharomyces cerevisiae trans-aconitate ...
|
|
64689
|
3-Isopropylmalate is the major endogenous substrate of the Saccharomyces cerevisiae trans-aconitate ...
|
|
64690
|
3-Isopropylmalate is the major endogenous substrate of the Saccharomyces cerevisiae trans-aconitate ...
|
|
64691
|
3-Isopropylmalate is the major endogenous substrate of the Saccharomyces cerevisiae trans-aconitate ...
|
|
64692
|
3-Isopropylmalate is the major endogenous substrate of the Saccharomyces cerevisiae trans-aconitate ...
|
|
64693
|
3-Isopropylmalate is the major endogenous substrate of the Saccharomyces cerevisiae trans-aconitate ...
|
|
64694
|
Identification of the gene and characterization of the activity of the trans-aconitate methyltransferase from ...
|
|
64695
|
Identification of the gene and characterization of the activity of the trans-aconitate methyltransferase from ...
|
|
64696
|
Identification of the gene and characterization of the activity of the trans-aconitate methyltransferase from ...
|
|
64697
|
Identification of the gene and characterization of the activity of the trans-aconitate methyltransferase from ...
|
|
64698
|
Identification of the gene and characterization of the activity of the trans-aconitate methyltransferase from ...
|
|
64699
|
Identification of the gene and characterization of the activity of the trans-aconitate methyltransferase from ...
|
|
64700
|
Identification of the gene and characterization of the activity of the trans-aconitate methyltransferase from ...
|
|
64701
|
Identification of the gene and characterization of the activity of the trans-aconitate methyltransferase from ...
|
|
64702
|
Identification of the gene and characterization of the activity of the trans-aconitate methyltransferase from ...
|
|
64703
|
Identification of the gene and characterization of the activity of the trans-aconitate methyltransferase from ...
|
|
64704
|
Identification of the gene and characterization of the activity of the trans-aconitate methyltransferase from ...
|
|
64705
|
Identification of the gene and characterization of the activity of the trans-aconitate methyltransferase from ...
|
|
64706
|
Identification of the gene and characterization of the activity of the trans-aconitate methyltransferase from ...
|
|
64707
|
Identification of the gene and characterization of the activity of the trans-aconitate methyltransferase from ...
|
|
64708
|
Identification of the gene and characterization of the activity of the trans-aconitate methyltransferase from ...
|
|
64709
|
Identification of the gene and characterization of the activity of the trans-aconitate methyltransferase from ...
|
|
64710
|
Identification of the gene and characterization of the activity of the trans-aconitate methyltransferase from ...
|
|
64711
|
Identification of the gene and characterization of the activity of the trans-aconitate methyltransferase from ...
|
|
64712
|
Identification of the gene and characterization of the activity of the trans-aconitate methyltransferase from ...
|
|
64713
|
Identification of the gene and characterization of the activity of the trans-aconitate methyltransferase from ...
|
|
64714
|
Identification of the gene and characterization of the activity of the trans-aconitate methyltransferase from ...
|
|
64715
|
Identification of the gene and characterization of the activity of the trans-aconitate methyltransferase from ...
|
|
64716
|
Identification of the gene and characterization of the activity of the trans-aconitate methyltransferase from ...
|
|
64717
|
Identification of the gene and characterization of the activity of the trans-aconitate methyltransferase from ...
|
|
64718
|
Reaction of the molybdenum- and copper-containing carbon monoxide dehydrogenase from Oligotropha ...
|
|
64719
|
Reaction of the molybdenum- and copper-containing carbon monoxide dehydrogenase from Oligotropha ...
|
|
64720
|
Reaction of the molybdenum- and copper-containing carbon monoxide dehydrogenase from Oligotropha ...
|
|
64721
|
Reaction of the molybdenum- and copper-containing carbon monoxide dehydrogenase from Oligotropha ...
|
|
64722
|
Reaction of the molybdenum- and copper-containing carbon monoxide dehydrogenase from Oligotropha ...
|
|
64723
|
Reaction of the molybdenum- and copper-containing carbon monoxide dehydrogenase from Oligotropha ...
|
|
64724
|
Reaction of the molybdenum- and copper-containing carbon monoxide dehydrogenase from Oligotropha ...
|
|
64725
|
Identification of a novel prostaglandin f(2alpha) synthase in Trypanosoma brucei.
|
|
64726
|
Identification of a novel prostaglandin f(2alpha) synthase in Trypanosoma brucei.
|
|
64727
|
Identification of a novel prostaglandin f(2alpha) synthase in Trypanosoma brucei.
|
|
64728
|
Identification of a novel prostaglandin f(2alpha) synthase in Trypanosoma brucei.
|
|
64729
|
Design, Synthesis and Biological Evaluation of Novel Peptide-Like Analogues as Selective COX-2 Inhibitors
|
|
64730
|
Design, Synthesis and Biological Evaluation of Novel Peptide-Like Analogues as Selective COX-2 Inhibitors
|
|
64731
|
Design, Synthesis and Biological Evaluation of Novel Peptide-Like Analogues as Selective COX-2 Inhibitors
|
|
64732
|
Design, Synthesis and Biological Evaluation of Novel Peptide-Like Analogues as Selective COX-2 Inhibitors
|
|
64733
|
Design, Synthesis and Biological Evaluation of Novel Peptide-Like Analogues as Selective COX-2 Inhibitors
|
|
64734
|
Design, Synthesis and Biological Evaluation of Novel Peptide-Like Analogues as Selective COX-2 Inhibitors
|
|
64735
|
Design, Synthesis and Biological Evaluation of Novel Peptide-Like Analogues as Selective COX-2 Inhibitors
|
|
64736
|
Design, Synthesis and Biological Evaluation of Novel Peptide-Like Analogues as Selective COX-2 Inhibitors
|
|
64737
|
Design, Synthesis and Biological Evaluation of Novel Peptide-Like Analogues as Selective COX-2 Inhibitors
|
|
64738
|
Design, Synthesis and Biological Evaluation of Novel Peptide-Like Analogues as Selective COX-2 Inhibitors
|
|
64739
|
Design, Synthesis and Biological Evaluation of Novel Peptide-Like Analogues as Selective COX-2 Inhibitors
|
|
64740
|
Design, Synthesis and Biological Evaluation of4-(Imidazolylmethyl)-2-(4-methylsulfonyl phenyl)-Quinoline ...
|
|
64741
|
Design, Synthesis and Biological Evaluation of4-(Imidazolylmethyl)-2-(4-methylsulfonyl phenyl)-Quinoline ...
|
|
64742
|
Design, Synthesis and Biological Evaluation of4-(Imidazolylmethyl)-2-(4-methylsulfonyl phenyl)-Quinoline ...
|
|
64743
|
Design, Synthesis and Biological Evaluation of4-(Imidazolylmethyl)-2-(4-methylsulfonyl phenyl)-Quinoline ...
|
|
64744
|
Design, Synthesis and Biological Evaluation of4-(Imidazolylmethyl)-2-(4-methylsulfonyl phenyl)-Quinoline ...
|
|
64745
|
Design, Synthesis and Biological Evaluation of4-(Imidazolylmethyl)-2-(4-methylsulfonyl phenyl)-Quinoline ...
|
|
64746
|
Design, Synthesis and Biological Evaluation of4-(Imidazolylmethyl)-2-(4-methylsulfonyl phenyl)-Quinoline ...
|
|
64747
|
Design, Synthesis and Biological Evaluation of4-(Imidazolylmethyl)-2-(4-methylsulfonyl phenyl)-Quinoline ...
|
|
64748
|
Design, Synthesis and Biological Evaluation of4-(Imidazolylmethyl)-2-(4-methylsulfonyl phenyl)-Quinoline ...
|
|
64749
|
Design, Synthesis and Biological Evaluation of4-(Imidazolylmethyl)-2-(4-methylsulfonyl phenyl)-Quinoline ...
|
|
64750
|
Design, Synthesis and Biological Evaluation of4-(Imidazolylmethyl)-2-(4-methylsulfonyl phenyl)-Quinoline ...
|
|
64751
|
Design, Synthesis and Biological Evaluation of4-(Imidazolylmethyl)-2-(4-methylsulfonyl phenyl)-Quinoline ...
|
|
64752
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64753
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64754
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64755
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64756
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64757
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64758
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64759
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64760
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64761
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64762
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64763
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64764
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64765
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64766
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64767
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64768
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64769
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64770
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64771
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64772
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64773
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64774
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64775
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64776
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64777
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64778
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64779
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64780
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64781
|
Design, synthesis and biological evaluation of 5-oxo-1,4,5,6,7,8 hexahydroquinoline derivatives as selective ...
|
|
64782
|
D-xylose degradation pathway in the halophilic archaeon Haloferax volcanii.
|
|
64783
|
D-xylose degradation pathway in the halophilic archaeon Haloferax volcanii.
|
|
64784
|
D-xylose degradation pathway in the halophilic archaeon Haloferax volcanii.
|
|
64785
|
D-xylose degradation pathway in the halophilic archaeon Haloferax volcanii.
|
|
64786
|
D-xylose degradation pathway in the halophilic archaeon Haloferax volcanii.
|
|
64787
|
D-xylose degradation pathway in the halophilic archaeon Haloferax volcanii.
|
|
64788
|
D-xylose degradation pathway in the halophilic archaeon Haloferax volcanii.
|
|
64789
|
D-xylose degradation pathway in the halophilic archaeon Haloferax volcanii.
|
|
64790
|
D-xylose degradation pathway in the halophilic archaeon Haloferax volcanii.
|
|
64791
|
D-xylose degradation pathway in the halophilic archaeon Haloferax volcanii.
|
|
64792
|
D-xylose degradation pathway in the halophilic archaeon Haloferax volcanii.
|
|
64793
|
D-xylose degradation pathway in the halophilic archaeon Haloferax volcanii.
|
|
64794
|
D-xylose degradation pathway in the halophilic archaeon Haloferax volcanii.
|
|
64795
|
D-xylose degradation pathway in the halophilic archaeon Haloferax volcanii.
|
|
64796
|
Active site and loop 4 movements within human glycolate oxidase: implications for substrate specificity and ...
|
|
64797
|
Active site and loop 4 movements within human glycolate oxidase: implications for substrate specificity and ...
|
|
64798
|
Active site and loop 4 movements within human glycolate oxidase: implications for substrate specificity and ...
|
|
64799
|
Crystal Structure of Aspirin-Acetylated Human Cyclooxygenase-2: Insight into the Formation of Products with ...
|
|
64800
|
Crystal Structure of Aspirin-Acetylated Human Cyclooxygenase-2: Insight into the Formation of Products with ...
|
|
64801
|
Crystal Structure of Aspirin-Acetylated Human Cyclooxygenase-2: Insight into the Formation of Products with ...
|
|
64802
|
Crystal Structure of Aspirin-Acetylated Human Cyclooxygenase-2: Insight into the Formation of Products with ...
|
|
64803
|
Crystal Structure of Aspirin-Acetylated Human Cyclooxygenase-2: Insight into the Formation of Products with ...
|
|
64804
|
Oxalate accumulation and regulation is independent of glycolate oxidase in rice leaves
|
|
64805
|
Oxalate accumulation and regulation is independent of glycolate oxidase in rice leaves
|
|
64806
|
Oxalate accumulation and regulation is independent of glycolate oxidase in rice leaves
|
|
64807
|
Oxalate accumulation and regulation is independent of glycolate oxidase in rice leaves
|
|
64808
|
Oxalate accumulation and regulation is independent of glycolate oxidase in rice leaves
|
|
64809
|
Oxalate accumulation and regulation is independent of glycolate oxidase in rice leaves
|
|
64810
|
Oxalate accumulation and regulation is independent of glycolate oxidase in rice leaves
|
|
64811
|
Oxalate accumulation and regulation is independent of glycolate oxidase in rice leaves
|
|
64812
|
Identification and characterization of VPO1, a new animal heme-containing peroxidase
|
|
64813
|
Molecular characterization of a novel type of prostamide/prostaglandin F synthase, belonging to the ...
|
|
64814
|
Molecular characterization of a novel type of prostamide/prostaglandin F synthase, belonging to the ...
|
|
64815
|
Molecular characterization of a novel type of prostamide/prostaglandin F synthase, belonging to the ...
|
|
64816
|
Molecular characterization of a novel type of prostamide/prostaglandin F synthase, belonging to the ...
|
|
64817
|
Molecular characterization of a novel type of prostamide/prostaglandin F synthase, belonging to the ...
|
|
64818
|
Molecular characterization of a novel type of prostamide/prostaglandin F synthase, belonging to the ...
|
|
64819
|
Molecular characterization of a novel type of prostamide/prostaglandin F synthase, belonging to the ...
|
|
64820
|
Purification and characterization of the Mycobacterium smegmatis catalase-peroxidase involved in isoniazid ...
|
|
64821
|
Purification and characterization of the Mycobacterium smegmatis catalase-peroxidase involved in isoniazid ...
|
|
64822
|
Purification and characterization of the Mycobacterium smegmatis catalase-peroxidase involved in isoniazid ...
|
|
64823
|
Purification and characterization of the Mycobacterium smegmatis catalase-peroxidase involved in isoniazid ...
|
|
64824
|
Purification and characterization of the Mycobacterium smegmatis catalase-peroxidase involved in isoniazid ...
|
|
64825
|
Purification and characterization of the Mycobacterium smegmatis catalase-peroxidase involved in isoniazid ...
|
|
64826
|
Purification and characterization of the Mycobacterium smegmatis catalase-peroxidase involved in isoniazid ...
|
|
64827
|
Purification and characterization of the Mycobacterium smegmatis catalase-peroxidase involved in isoniazid ...
|
|
64828
|
Purification and characterization of the Mycobacterium smegmatis catalase-peroxidase involved in isoniazid ...
|
|
64829
|
Purification and characterization of the Mycobacterium smegmatis catalase-peroxidase involved in isoniazid ...
|
|
64830
|
Purification and characterization of the Mycobacterium smegmatis catalase-peroxidase involved in isoniazid ...
|
|
64831
|
Purification and characterization of the Mycobacterium smegmatis catalase-peroxidase involved in isoniazid ...
|
|
64832
|
Purification and characterization of the Mycobacterium smegmatis catalase-peroxidase involved in isoniazid ...
|
|
64833
|
Characterization of patient mutations in human persulfide dioxygenase (ETHE1) involved in H2S catabolism.
|
|
64834
|
Characterization of patient mutations in human persulfide dioxygenase (ETHE1) involved in H2S catabolism.
|
|
64835
|
Characterization of patient mutations in human persulfide dioxygenase (ETHE1) involved in H2S catabolism.
|
|
64836
|
Characterization of patient mutations in human persulfide dioxygenase (ETHE1) involved in H2S catabolism.
|
|
64837
|
Purification and characterization of recombinant histidine-tagged human platelet 12-lipoxygenase expressed in ...
|
|
64838
|
Purification and characterization of recombinant histidine-tagged human platelet 12-lipoxygenase expressed in ...
|
|
64839
|
Lipoxin synthase activity of human platelet 12-lipoxygenase
|
|
64840
|
Lipoxin synthase activity of human platelet 12-lipoxygenase
|
|
64841
|
Lipoxin synthase activity of human platelet 12-lipoxygenase
|
|
64842
|
Novel genes encoding 2-aminophenol 1,6-dioxygenase from Pseudomonas species AP-3 growing on 2-aminophenol and ...
|
|
64843
|
Novel genes encoding 2-aminophenol 1,6-dioxygenase from Pseudomonas species AP-3 growing on 2-aminophenol and ...
|
|
64844
|
Novel genes encoding 2-aminophenol 1,6-dioxygenase from Pseudomonas species AP-3 growing on 2-aminophenol and ...
|
|
64845
|
Novel genes encoding 2-aminophenol 1,6-dioxygenase from Pseudomonas species AP-3 growing on 2-aminophenol and ...
|
|
64846
|
Novel genes encoding 2-aminophenol 1,6-dioxygenase from Pseudomonas species AP-3 growing on 2-aminophenol and ...
|
|
64847
|
Novel genes encoding 2-aminophenol 1,6-dioxygenase from Pseudomonas species AP-3 growing on 2-aminophenol and ...
|
|
64848
|
Novel genes encoding 2-aminophenol 1,6-dioxygenase from Pseudomonas species AP-3 growing on 2-aminophenol and ...
|
|
64849
|
Novel genes encoding 2-aminophenol 1,6-dioxygenase from Pseudomonas species AP-3 growing on 2-aminophenol and ...
|
|
64850
|
Novel genes encoding 2-aminophenol 1,6-dioxygenase from Pseudomonas species AP-3 growing on 2-aminophenol and ...
|
|
64851
|
Novel genes encoding 2-aminophenol 1,6-dioxygenase from Pseudomonas species AP-3 growing on 2-aminophenol and ...
|
|
64852
|
nag genes of Ralstonia (formerly Pseudomonas) sp. strain U2 encoding enzymes for gentisate catabolism.
|
|
64853
|
nag genes of Ralstonia (formerly Pseudomonas) sp. strain U2 encoding enzymes for gentisate catabolism.
|
|
64854
|
nag genes of Ralstonia (formerly Pseudomonas) sp. strain U2 encoding enzymes for gentisate catabolism.
|
|
64855
|
Deciphering the genetic determinants for aerobic nicotinic acid degradation: the nic cluster from Pseudomonas ...
|
|
64856
|
Deciphering the genetic determinants for aerobic nicotinic acid degradation: the nic cluster from Pseudomonas ...
|
|
64857
|
Deciphering the genetic determinants for aerobic nicotinic acid degradation: the nic cluster from Pseudomonas ...
|
|
64858
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64859
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64860
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64861
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64862
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64863
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64864
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64865
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64866
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64867
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64868
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64869
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64870
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64871
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64872
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64873
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64874
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64875
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64876
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64877
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64878
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64879
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64880
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64881
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64882
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64883
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64884
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64885
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64886
|
Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response
|
|
64887
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64888
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64889
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64890
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64891
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64892
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64893
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64894
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64895
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64896
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64897
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64898
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64899
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64900
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64901
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64902
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64903
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64904
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64905
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64906
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64907
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64908
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64909
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64910
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64911
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64912
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64913
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64914
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64915
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64916
|
Exploring sponge-derived terpenoids for their potency and selectivity against 12-human, 15-human, and ...
|
|
64917
|
A novel biotin protein required for reductive carboxylation of 2-oxoglutarate by isocitrate dehydrogenase in ...
|
|
64918
|
A novel biotin protein required for reductive carboxylation of 2-oxoglutarate by isocitrate dehydrogenase in ...
|
|
64919
|
Point mutation (R153H or R153C) in Escherichia coli isocitrate dehydrogenase: Biochemical characterization and ...
|
|
64920
|
Point mutation (R153H or R153C) in Escherichia coli isocitrate dehydrogenase: Biochemical characterization and ...
|
|
64921
|
Point mutation (R153H or R153C) in Escherichia coli isocitrate dehydrogenase: Biochemical characterization and ...
|
|
64922
|
Point mutation (R153H or R153C) in Escherichia coli isocitrate dehydrogenase: Biochemical characterization and ...
|
|
64923
|
Point mutation (R153H or R153C) in Escherichia coli isocitrate dehydrogenase: Biochemical characterization and ...
|
|
64924
|
Structural, kinetic and chemical mechanism of isocitrate dehydrogenase-1 from Mycobacterium tuberculosis
|
|
64925
|
Structural, kinetic and chemical mechanism of isocitrate dehydrogenase-1 from Mycobacterium tuberculosis
|
|
64926
|
Structural, kinetic and chemical mechanism of isocitrate dehydrogenase-1 from Mycobacterium tuberculosis
|
|
64927
|
Structural, kinetic and chemical mechanism of isocitrate dehydrogenase-1 from Mycobacterium tuberculosis
|
|
64928
|
Structural, kinetic and chemical mechanism of isocitrate dehydrogenase-1 from Mycobacterium tuberculosis
|
|
64929
|
Structural, kinetic and chemical mechanism of isocitrate dehydrogenase-1 from Mycobacterium tuberculosis
|
|
64930
|
Structural, kinetic and chemical mechanism of isocitrate dehydrogenase-1 from Mycobacterium tuberculosis
|
|
64931
|
Structural, kinetic and chemical mechanism of isocitrate dehydrogenase-1 from Mycobacterium tuberculosis
|
|
64932
|
A highly active decarboxylating dehydrogenase with rationally inverted coenzyme specificity.
|
|
64933
|
A highly active decarboxylating dehydrogenase with rationally inverted coenzyme specificity.
|
|
64934
|
A highly active decarboxylating dehydrogenase with rationally inverted coenzyme specificity.
|
|
64935
|
A highly active decarboxylating dehydrogenase with rationally inverted coenzyme specificity.
|
|
64936
|
A highly active decarboxylating dehydrogenase with rationally inverted coenzyme specificity.
|
|
64937
|
A highly active decarboxylating dehydrogenase with rationally inverted coenzyme specificity.
|
|
64938
|
A highly active decarboxylating dehydrogenase with rationally inverted coenzyme specificity.
|
|
64939
|
A highly active decarboxylating dehydrogenase with rationally inverted coenzyme specificity.
|
|
64940
|
A highly active decarboxylating dehydrogenase with rationally inverted coenzyme specificity.
|
|
64941
|
A highly active decarboxylating dehydrogenase with rationally inverted coenzyme specificity.
|
|
64942
|
A highly active decarboxylating dehydrogenase with rationally inverted coenzyme specificity.
|
|
64943
|
A highly active decarboxylating dehydrogenase with rationally inverted coenzyme specificity.
|
|
64944
|
A highly active decarboxylating dehydrogenase with rationally inverted coenzyme specificity.
|
|
64945
|
A highly active decarboxylating dehydrogenase with rationally inverted coenzyme specificity.
|
|
64946
|
A highly active decarboxylating dehydrogenase with rationally inverted coenzyme specificity.
|
|
64947
|
A highly active decarboxylating dehydrogenase with rationally inverted coenzyme specificity.
|
|
64948
|
A highly active decarboxylating dehydrogenase with rationally inverted coenzyme specificity.
|
|
64949
|
Enzymatic characterization of a monomeric isocitrate dehydrogenase from Streptomyces lividans TK54
|
|
64950
|
Enzymatic characterization of a monomeric isocitrate dehydrogenase from Streptomyces lividans TK54
|
|
64951
|
Enzymatic characterization of a monomeric isocitrate dehydrogenase from Streptomyces lividans TK54
|
|
64952
|
Enzymatic characterization of a monomeric isocitrate dehydrogenase from Streptomyces lividans TK54
|
|
64953
|
Biochemical characteristics and inhibitor selectivity of mouse indoleamine 2,3-dioxygenase-2.
|
|
64954
|
Biochemical characteristics and inhibitor selectivity of mouse indoleamine 2,3-dioxygenase-2.
|
|
64955
|
Biochemical characteristics and inhibitor selectivity of mouse indoleamine 2,3-dioxygenase-2.
|
|
64956
|
Biochemical characteristics and inhibitor selectivity of mouse indoleamine 2,3-dioxygenase-2.
|
|
64957
|
Biochemical characteristics and inhibitor selectivity of mouse indoleamine 2,3-dioxygenase-2.
|
|
64958
|
Biochemical characteristics and inhibitor selectivity of mouse indoleamine 2,3-dioxygenase-2.
|
|
64959
|
Biochemical characteristics and inhibitor selectivity of mouse indoleamine 2,3-dioxygenase-2.
|
|
64960
|
Biochemical characteristics and inhibitor selectivity of mouse indoleamine 2,3-dioxygenase-2.
|
|
64961
|
Biochemical characteristics and inhibitor selectivity of mouse indoleamine 2,3-dioxygenase-2.
|
|
64962
|
Biochemical characteristics and inhibitor selectivity of mouse indoleamine 2,3-dioxygenase-2.
|
|
64963
|
Biochemical characteristics and inhibitor selectivity of mouse indoleamine 2,3-dioxygenase-2.
|
|
64964
|
Biochemical characteristics and inhibitor selectivity of mouse indoleamine 2,3-dioxygenase-2.
|
|
64965
|
Biochemical characteristics and inhibitor selectivity of mouse indoleamine 2,3-dioxygenase-2.
|
|
64966
|
Purification, characterization, and mechanism of a flavin mononucleotide-dependent 2-nitropropane dioxygenase ...
|
|
64967
|
Purification, characterization, and mechanism of a flavin mononucleotide-dependent 2-nitropropane dioxygenase ...
|
|
64968
|
Purification, characterization, and mechanism of a flavin mononucleotide-dependent 2-nitropropane dioxygenase ...
|
|
64969
|
Purification, characterization, and mechanism of a flavin mononucleotide-dependent 2-nitropropane dioxygenase ...
|
|
64970
|
Purification, characterization, and mechanism of a flavin mononucleotide-dependent 2-nitropropane dioxygenase ...
|
|
64971
|
Purification, characterization, and mechanism of a flavin mononucleotide-dependent 2-nitropropane dioxygenase ...
|
|
64972
|
Catechol dioxygenases from Escherichia coli (MhpB) and Alcaligenes eutrophus (MpcI): sequence analysis and ...
|
|
64973
|
Catechol dioxygenases from Escherichia coli (MhpB) and Alcaligenes eutrophus (MpcI): sequence analysis and ...
|
|
64974
|
Catechol dioxygenases from Escherichia coli (MhpB) and Alcaligenes eutrophus (MpcI): sequence analysis and ...
|
|
64975
|
Catechol dioxygenases from Escherichia coli (MhpB) and Alcaligenes eutrophus (MpcI): sequence analysis and ...
|
|
64976
|
Catechol dioxygenases from Escherichia coli (MhpB) and Alcaligenes eutrophus (MpcI): sequence analysis and ...
|
|
64977
|
Catechol dioxygenases from Escherichia coli (MhpB) and Alcaligenes eutrophus (MpcI): sequence analysis and ...
|
|
64978
|
Catechol dioxygenases from Escherichia coli (MhpB) and Alcaligenes eutrophus (MpcI): sequence analysis and ...
|
|
64979
|
Catechol dioxygenases from Escherichia coli (MhpB) and Alcaligenes eutrophus (MpcI): sequence analysis and ...
|
|
64980
|
Catechol dioxygenases from Escherichia coli (MhpB) and Alcaligenes eutrophus (MpcI): sequence analysis and ...
|
|
64981
|
Catechol dioxygenases from Escherichia coli (MhpB) and Alcaligenes eutrophus (MpcI): sequence analysis and ...
|
|
64982
|
Biochemical and molecular characterization of the NAD(+)-dependent isocitrate dehydrogenase from the ...
|
|
64983
|
Biochemical and molecular characterization of the NAD(+)-dependent isocitrate dehydrogenase from the ...
|
|
64984
|
Purification and enzymatic properties of peptide:N-glycanase from C3H mouse-derived L-929 fibroblast cells. ...
|
|
64985
|
The ureide-degrading reactions of purine ring catabolism employ three amidohydrolases and one aminohydrolase ...
|
|
64986
|
The ureide-degrading reactions of purine ring catabolism employ three amidohydrolases and one aminohydrolase ...
|
|
64987
|
The ureide-degrading reactions of purine ring catabolism employ three amidohydrolases and one aminohydrolase ...
|
|
64988
|
The ureide-degrading reactions of purine ring catabolism employ three amidohydrolases and one aminohydrolase ...
|
|
64989
|
The ureide-degrading reactions of purine ring catabolism employ three amidohydrolases and one aminohydrolase ...
|
|
64990
|
The ureide-degrading reactions of purine ring catabolism employ three amidohydrolases and one aminohydrolase ...
|
|
64991
|
The ureide-degrading reactions of purine ring catabolism employ three amidohydrolases and one aminohydrolase ...
|
|
64992
|
The ureide-degrading reactions of purine ring catabolism employ three amidohydrolases and one aminohydrolase ...
|
|
64993
|
The ureide-degrading reactions of purine ring catabolism employ three amidohydrolases and one aminohydrolase ...
|
|
64994
|
The ureide-degrading reactions of purine ring catabolism employ three amidohydrolases and one aminohydrolase ...
|
|
64995
|
Identification and characterization of proteins involved in rice urea and arginine catabolism
|
|
64996
|
Identification and characterization of proteins involved in rice urea and arginine catabolism
|
|
64997
|
Identification and characterization of proteins involved in rice urea and arginine catabolism
|
|
64998
|
Molecular identification of a functional homologue of the mammalian fatty acid amide hydrolase in Arabidopsis ...
|
|
64999
|
Molecular identification of a functional homologue of the mammalian fatty acid amide hydrolase in Arabidopsis ...
|
|
65000
|
Molecular identification of a functional homologue of the mammalian fatty acid amide hydrolase in Arabidopsis ...
|





 If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.
If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.