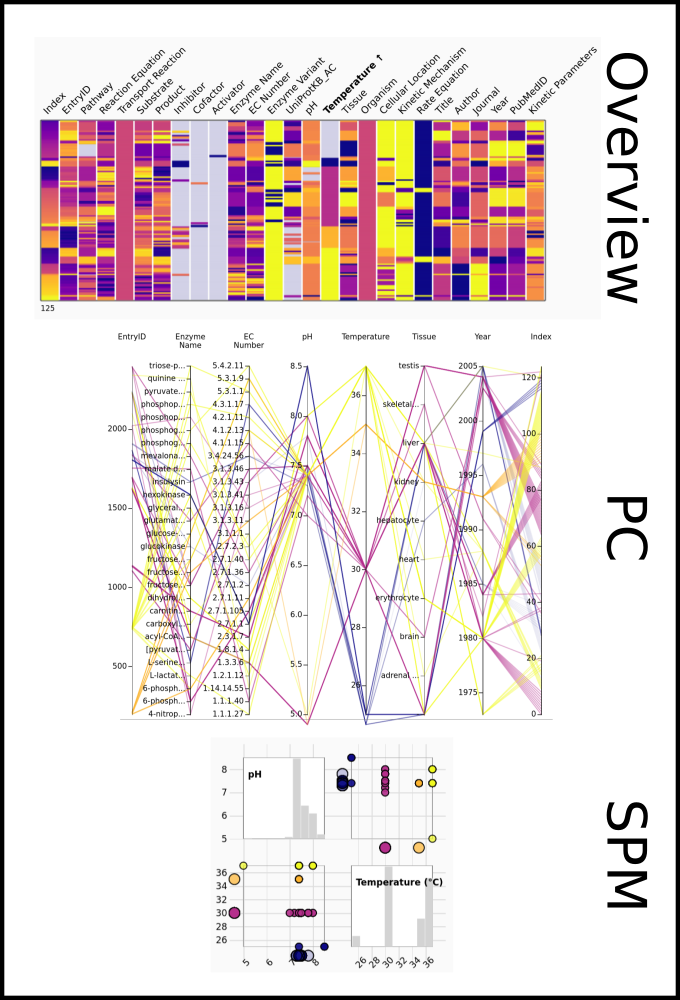
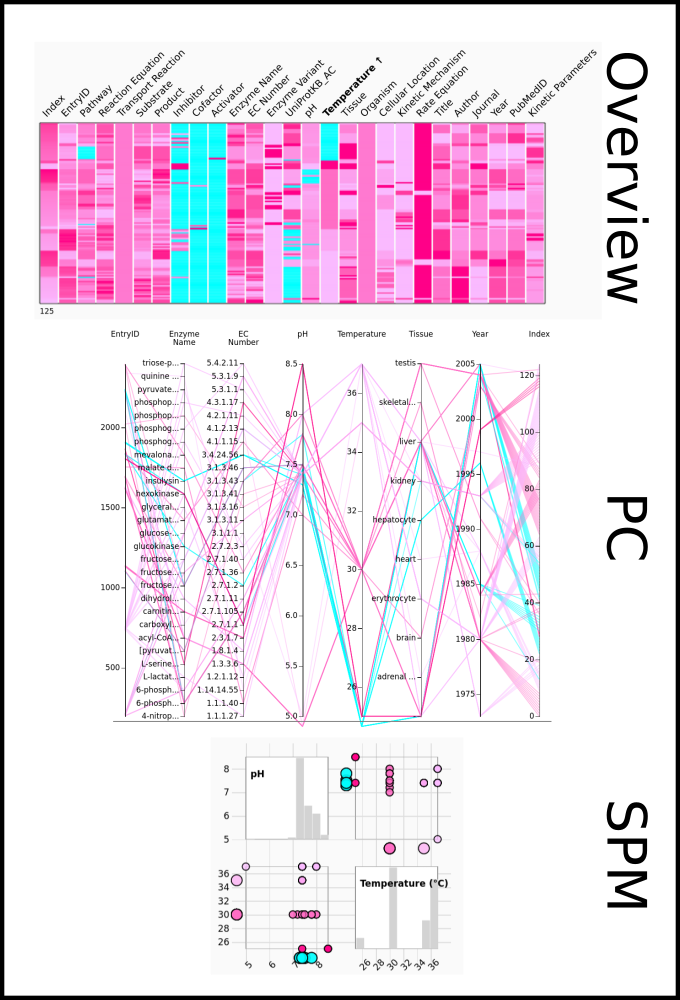
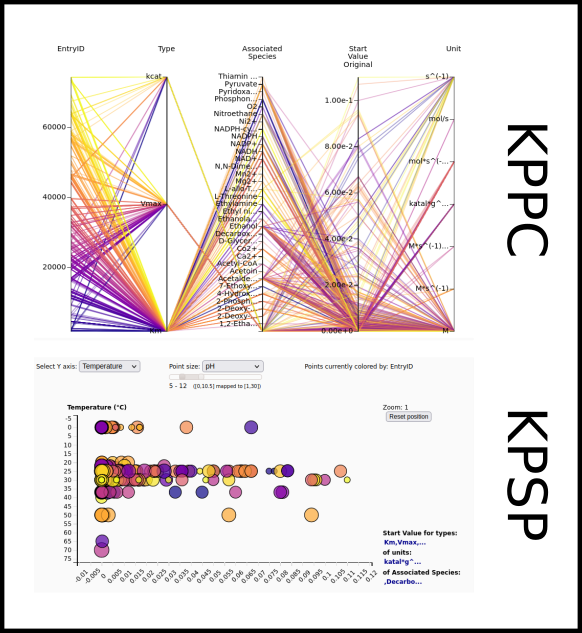
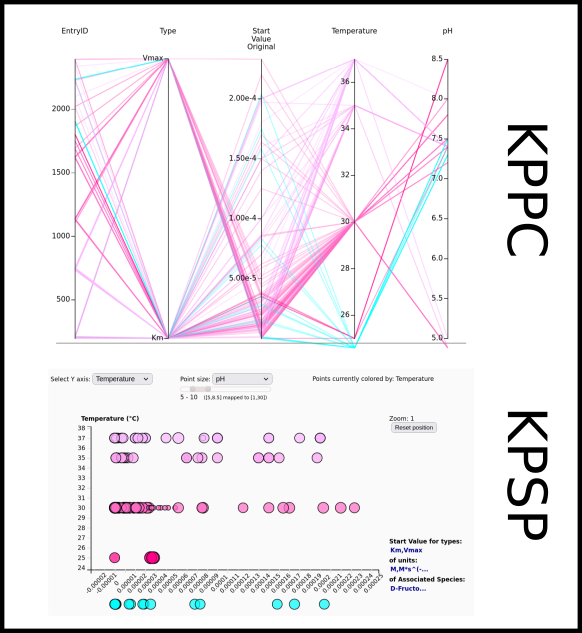
To improve SABIO-RK data interoperability, semantic markup was added to web pages as described and defined by
This structured information makes it easier to discover, collate and analyse our data.
|
13008
|
Purification and properties of cystathionine beta-synthase from human liver. Evidence for identical subunits
|
|
13009
|
Purification and properties of cystathionine beta-synthase from human liver. Evidence for identical subunits
|
|
13010
|
Purification and characterization of avian liver 3-hydroxy-3-methylglutaryl coenzyme A lyase
|
|
13011
|
Comparison of the D-glutamate-adding enzymes from selected gram-positive and gram-negative bacteria
|
|
13012
|
Comparison of the D-glutamate-adding enzymes from selected gram-positive and gram-negative bacteria
|
|
13013
|
Comparison of the D-glutamate-adding enzymes from selected gram-positive and gram-negative bacteria
|
|
13014
|
Comparison of the D-glutamate-adding enzymes from selected gram-positive and gram-negative bacteria
|
|
13015
|
Comparison of the D-glutamate-adding enzymes from selected gram-positive and gram-negative bacteria
|
|
13016
|
Comparison of the D-glutamate-adding enzymes from selected gram-positive and gram-negative bacteria
|
|
13017
|
Comparison of the D-glutamate-adding enzymes from selected gram-positive and gram-negative bacteria
|
|
13018
|
Comparison of the D-glutamate-adding enzymes from selected gram-positive and gram-negative bacteria
|
|
13019
|
Comparison of the D-glutamate-adding enzymes from selected gram-positive and gram-negative bacteria
|
|
13020
|
Comparison of the D-glutamate-adding enzymes from selected gram-positive and gram-negative bacteria
|
|
13021
|
Comparison of the D-glutamate-adding enzymes from selected gram-positive and gram-negative bacteria
|
|
13022
|
Comparison of the D-glutamate-adding enzymes from selected gram-positive and gram-negative bacteria
|
|
13023
|
Comparison of the D-glutamate-adding enzymes from selected gram-positive and gram-negative bacteria
|
|
13024
|
Comparison of the D-glutamate-adding enzymes from selected gram-positive and gram-negative bacteria
|
|
13025
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13026
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13027
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13028
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13029
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13030
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13031
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13032
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13033
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13034
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13035
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13036
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13037
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13038
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13039
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13040
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13041
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13042
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13043
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13044
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13045
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13046
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13047
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13048
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13049
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13050
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13051
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13052
|
Kinetic and mutation analyses of aspartate kinase from Thermus flavus
|
|
13053
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13054
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13055
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13056
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13057
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13058
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13059
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13060
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13061
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13062
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13063
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13064
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13065
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13066
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13067
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13068
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13069
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13070
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13071
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13072
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13073
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13074
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13075
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13076
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13077
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13078
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13079
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13080
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13081
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13082
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13083
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13084
|
Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase ...
|
|
13085
|
Identification of catalytic residues of Ca2+-independent 1,2-alpha-D-mannosidase from Aspergillus saitoi by ...
|
|
13086
|
Identification of catalytic residues of Ca2+-independent 1,2-alpha-D-mannosidase from Aspergillus saitoi by ...
|
|
13087
|
Identification of catalytic residues of Ca2+-independent 1,2-alpha-D-mannosidase from Aspergillus saitoi by ...
|
|
13088
|
Identification of catalytic residues of Ca2+-independent 1,2-alpha-D-mannosidase from Aspergillus saitoi by ...
|
|
13089
|
Identification of catalytic residues of Ca2+-independent 1,2-alpha-D-mannosidase from Aspergillus saitoi by ...
|
|
13090
|
Identification of catalytic residues of Ca2+-independent 1,2-alpha-D-mannosidase from Aspergillus saitoi by ...
|
|
13091
|
Identification of catalytic residues of Ca2+-independent 1,2-alpha-D-mannosidase from Aspergillus saitoi by ...
|
|
13092
|
Identification of catalytic residues of Ca2+-independent 1,2-alpha-D-mannosidase from Aspergillus saitoi by ...
|
|
13093
|
Identification of catalytic residues of Ca2+-independent 1,2-alpha-D-mannosidase from Aspergillus saitoi by ...
|
|
13094
|
Identification of catalytic residues of Ca2+-independent 1,2-alpha-D-mannosidase from Aspergillus saitoi by ...
|
|
13095
|
Purification and characterization of the alanine aminotransferase from the hyperthermophilic Archaeon ...
|
|
13096
|
Purification and characterization of the alanine aminotransferase from the hyperthermophilic Archaeon ...
|
|
13097
|
Purification and characterization of the alanine aminotransferase from the hyperthermophilic Archaeon ...
|
|
13098
|
Purification and characterization of the alanine aminotransferase from the hyperthermophilic Archaeon ...
|
|
13099
|
Purification and characterization of the alanine aminotransferase from the hyperthermophilic Archaeon ...
|
|
13100
|
Purification and characterization of the alanine aminotransferase from the hyperthermophilic Archaeon ...
|
|
13101
|
Purification and characterization of the alanine aminotransferase from the hyperthermophilic Archaeon ...
|
|
13102
|
Purification and characterization of the alanine aminotransferase from the hyperthermophilic Archaeon ...
|
|
13103
|
Mechanism for regulating the distribution of glucose carbon between the Embden-Meyerhof and ...
|
|
13104
|
Mechanism for regulating the distribution of glucose carbon between the Embden-Meyerhof and ...
|
|
13105
|
Mechanism for regulating the distribution of glucose carbon between the Embden-Meyerhof and ...
|
|
13106
|
Mechanism for regulating the distribution of glucose carbon between the Embden-Meyerhof and ...
|
|
13107
|
Mechanism for regulating the distribution of glucose carbon between the Embden-Meyerhof and ...
|
|
13108
|
Mechanism for regulating the distribution of glucose carbon between the Embden-Meyerhof and ...
|
|
13109
|
Differences in catalytic properties between cerebral cytoplasmic and mitochondrial hexokinases
|
|
13110
|
Differences in catalytic properties between cerebral cytoplasmic and mitochondrial hexokinases
|
|
13111
|
Differences in catalytic properties between cerebral cytoplasmic and mitochondrial hexokinases
|
|
13112
|
Differences in catalytic properties between cerebral cytoplasmic and mitochondrial hexokinases
|
|
13113
|
Differences in catalytic properties between cerebral cytoplasmic and mitochondrial hexokinases
|
|
13114
|
Differences in catalytic properties between cerebral cytoplasmic and mitochondrial hexokinases
|
|
13115
|
Differences in catalytic properties between cerebral cytoplasmic and mitochondrial hexokinases
|
|
13116
|
Differences in catalytic properties between cerebral cytoplasmic and mitochondrial hexokinases
|
|
13117
|
The purification and properties of human liver ketohexokinase. A role for ketohexokinase and ...
|
|
13118
|
The purification and properties of human liver ketohexokinase. A role for ketohexokinase and ...
|
|
13119
|
The purification and properties of human liver ketohexokinase. A role for ketohexokinase and ...
|
|
13120
|
The purification and properties of human liver ketohexokinase. A role for ketohexokinase and ...
|
|
13121
|
The purification and properties of human liver ketohexokinase. A role for ketohexokinase and ...
|
|
13122
|
Isolation and characterization of dihydropteridine reductase from human liver
|
|
13123
|
Isolation and characterization of dihydropteridine reductase from human liver
|
|
13124
|
Isolation and characterization of dihydropteridine reductase from human liver
|
|
13125
|
Isolation and characterization of dihydropteridine reductase from human liver
|
|
13126
|
Streptococcus faecalis aspartate transcarbamylase. I. Purification and general properties
|
|
13127
|
Streptococcus faecalis aspartate transcarbamylase. I. Purification and general properties
|
|
13128
|
Aspartate transcarbamylase from Streptococcus faecalis. Purification, properties, and nature of an allosteric ...
|
|
13129
|
Aspartate transcarbamylase from Streptococcus faecalis. Purification, properties, and nature of an allosteric ...
|
|
13130
|
Activation of D-aspartic acid for incorporation into peptidoglycan
|
|
13131
|
Activation of D-aspartic acid for incorporation into peptidoglycan
|
|
13132
|
The hyperthermophilic glycolytic enzyme enolase in the archaeon, Pyrococcus furiosus: comparison with ...
|
|
13133
|
The hyperthermophilic glycolytic enzyme enolase in the archaeon, Pyrococcus furiosus: comparison with ...
|
|
13134
|
The hyperthermophilic glycolytic enzyme enolase in the archaeon, Pyrococcus furiosus: comparison with ...
|
|
13135
|
The hyperthermophilic glycolytic enzyme enolase in the archaeon, Pyrococcus furiosus: comparison with ...
|
|
13136
|
The hyperthermophilic glycolytic enzyme enolase in the archaeon, Pyrococcus furiosus: comparison with ...
|
|
13137
|
The hyperthermophilic glycolytic enzyme enolase in the archaeon, Pyrococcus furiosus: comparison with ...
|
|
13138
|
Purification and lipid dependence of the recombinant hyaluronan synthases from Streptococcus pyogenes and ...
|
|
13139
|
Purification and lipid dependence of the recombinant hyaluronan synthases from Streptococcus pyogenes and ...
|
|
13140
|
Purification and lipid dependence of the recombinant hyaluronan synthases from Streptococcus pyogenes and ...
|
|
13141
|
Purification and lipid dependence of the recombinant hyaluronan synthases from Streptococcus pyogenes and ...
|
|
13142
|
Purification and lipid dependence of the recombinant hyaluronan synthases from Streptococcus pyogenes and ...
|
|
13143
|
Purification and lipid dependence of the recombinant hyaluronan synthases from Streptococcus pyogenes and ...
|
|
13144
|
Purification and lipid dependence of the recombinant hyaluronan synthases from Streptococcus pyogenes and ...
|
|
13145
|
Purification and lipid dependence of the recombinant hyaluronan synthases from Streptococcus pyogenes and ...
|
|
13146
|
D-glyceraldehyde 3-phosphate dehydrogenases of higher plants
|
|
13147
|
D-glyceraldehyde 3-phosphate dehydrogenases of higher plants
|
|
13148
|
D-glyceraldehyde 3-phosphate dehydrogenases of higher plants
|
|
13149
|
D-glyceraldehyde 3-phosphate dehydrogenases of higher plants
|
|
13150
|
D-glyceraldehyde 3-phosphate dehydrogenases of higher plants
|
|
13151
|
D-glyceraldehyde 3-phosphate dehydrogenases of higher plants
|
|
13152
|
D-glyceraldehyde 3-phosphate dehydrogenases of higher plants
|
|
13153
|
D-glyceraldehyde 3-phosphate dehydrogenases of higher plants
|
|
13154
|
D-glyceraldehyde 3-phosphate dehydrogenases of higher plants
|
|
13155
|
D-glyceraldehyde 3-phosphate dehydrogenases of higher plants
|
|
13156
|
D-glyceraldehyde 3-phosphate dehydrogenases of higher plants
|
|
13157
|
D-glyceraldehyde 3-phosphate dehydrogenases of higher plants
|
|
13158
|
D-glyceraldehyde 3-phosphate dehydrogenases of higher plants
|
|
13159
|
D-glyceraldehyde 3-phosphate dehydrogenases of higher plants
|
|
13160
|
D-glyceraldehyde 3-phosphate dehydrogenases of higher plants
|
|
13161
|
Divalent cation and pH dependent primary isotope effects in the enolase reaction
|
|
13162
|
Divalent cation and pH dependent primary isotope effects in the enolase reaction
|
|
13163
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13164
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13165
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13166
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13167
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13168
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13169
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13170
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13171
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13172
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13173
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13174
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13175
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13176
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13177
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13178
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13179
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13180
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13181
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13182
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13183
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13184
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13185
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13186
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13187
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13188
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13189
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13190
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13191
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13192
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13193
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13194
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13195
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13196
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13197
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13198
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13199
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13200
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13201
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13202
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13203
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13204
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13205
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13206
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13207
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13208
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13209
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13210
|
Kinetic properties of human liver alcohol dehydrogenase: oxidation of alcohols by class I isoenzymes
|
|
13211
|
The N-terminus of Drosophila SU(VAR)3-9 mediates dimerization and regulates its methyltransferase activity
|
|
13212
|
The N-terminus of Drosophila SU(VAR)3-9 mediates dimerization and regulates its methyltransferase activity
|
|
13213
|
Interactions between beta-lactam antibiotics and isolated membranes of Streptococcus faecalis ATCC 9790
|
|
13214
|
Interactions between beta-lactam antibiotics and isolated membranes of Streptococcus faecalis ATCC 9790
|
|
13215
|
Interactions between beta-lactam antibiotics and isolated membranes of Streptococcus faecalis ATCC 9790
|
|
13216
|
Interactions between beta-lactam antibiotics and isolated membranes of Streptococcus faecalis ATCC 9790
|
|
13217
|
Interactions between beta-lactam antibiotics and isolated membranes of Streptococcus faecalis ATCC 9790
|
|
13218
|
Interactions between beta-lactam antibiotics and isolated membranes of Streptococcus faecalis ATCC 9790
|
|
13219
|
Interactions between beta-lactam antibiotics and isolated membranes of Streptococcus faecalis ATCC 9790
|
|
13220
|
Interactions between beta-lactam antibiotics and isolated membranes of Streptococcus faecalis ATCC 9790
|
|
13221
|
Interactions between beta-lactam antibiotics and isolated membranes of Streptococcus faecalis ATCC 9790
|
|
13222
|
Interactions between beta-lactam antibiotics and isolated membranes of Streptococcus faecalis ATCC 9790
|
|
13223
|
Interactions between beta-lactam antibiotics and isolated membranes of Streptococcus faecalis ATCC 9790
|
|
13224
|
Interactions between beta-lactam antibiotics and isolated membranes of Streptococcus faecalis ATCC 9790
|
|
13225
|
Interactions between beta-lactam antibiotics and isolated membranes of Streptococcus faecalis ATCC 9790
|
|
13226
|
Interactions between beta-lactam antibiotics and isolated membranes of Streptococcus faecalis ATCC 9790
|
|
13227
|
The exchange reaction of peptides R-D-alanyl-D-alanine with D-[14C]alanine to R-D-alanyl-D-[14C]alanine and ...
|
|
13228
|
The exchange reaction of peptides R-D-alanyl-D-alanine with D-[14C]alanine to R-D-alanyl-D-[14C]alanine and ...
|
|
13229
|
Mechanistic Characterization of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa
|
|
13230
|
Mechanistic Characterization of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa
|
|
13231
|
Mechanistic Characterization of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa
|
|
13232
|
Mechanistic Characterization of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa
|
|
13233
|
Mechanistic Characterization of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa
|
|
13234
|
Mechanistic Characterization of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa
|
|
13235
|
Mechanistic Characterization of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa
|
|
13236
|
Mechanistic Characterization of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa
|
|
13237
|
Mechanistic Characterization of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa
|
|
13238
|
Mechanistic Characterization of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa
|
|
13239
|
Mechanistic Characterization of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa
|
|
13240
|
Mechanistic Characterization of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa
|
|
13241
|
Mechanistic Characterization of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa
|
|
13242
|
Mechanistic Characterization of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa
|
|
13243
|
Mechanistic Characterization of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa
|
|
13244
|
Mechanistic Characterization of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa
|
|
13245
|
Mechanistic Characterization of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa
|
|
13246
|
Mechanistic Characterization of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa
|
|
13247
|
Mechanistic Characterization of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa
|
|
13248
|
Mechanistic Characterization of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa
|
|
13249
|
Mechanistic Characterization of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa
|
|
13250
|
Mechanistic Characterization of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa
|
|
13251
|
Mechanistic Characterization of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa
|
|
13252
|
Mechanistic Characterization of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa
|
|
13253
|
Saccharomyces cerevisiae GNA1, an Essential Gene Encoding a Novel Acetyltransferase Involved in ...
|
|
13254
|
Saccharomyces cerevisiae GNA1, an Essential Gene Encoding a Novel Acetyltransferase Involved in ...
|
|
13255
|
Saccharomyces cerevisiae GNA1, an Essential Gene Encoding a Novel Acetyltransferase Involved in ...
|
|
13256
|
Saccharomyces cerevisiae GNA1, an Essential Gene Encoding a Novel Acetyltransferase Involved in ...
|
|
13257
|
Saccharomyces cerevisiae GNA1, an Essential Gene Encoding a Novel Acetyltransferase Involved in ...
|
|
13258
|
Saccharomyces cerevisiae GNA1, an Essential Gene Encoding a Novel Acetyltransferase Involved in ...
|
|
13259
|
Saccharomyces cerevisiae GNA1, an Essential Gene Encoding a Novel Acetyltransferase Involved in ...
|
|
13260
|
Saccharomyces cerevisiae GNA1, an Essential Gene Encoding a Novel Acetyltransferase Involved in ...
|
|
13261
|
Saccharomyces cerevisiae GNA1, an Essential Gene Encoding a Novel Acetyltransferase Involved in ...
|
|
13262
|
Saccharomyces cerevisiae GNA1, an Essential Gene Encoding a Novel Acetyltransferase Involved in ...
|
|
13263
|
Saccharomyces cerevisiae GNA1, an Essential Gene Encoding a Novel Acetyltransferase Involved in ...
|
|
13264
|
Magnesium-dependent ATPase activity and cooperativity of magnesium chelatase from Synechocystis sp. PCC6803
|
|
13265
|
Magnesium-dependent ATPase activity and cooperativity of magnesium chelatase from Synechocystis sp. PCC6803
|
|
13266
|
Magnesium-dependent ATPase activity and cooperativity of magnesium chelatase from Synechocystis sp. PCC6803
|
|
13267
|
Catalytic mechanism of type 2 isopentenyl diphosphate:dimethylallyl diphosphate isomerase: verification of a ...
|
|
13268
|
Catalytic mechanism of type 2 isopentenyl diphosphate:dimethylallyl diphosphate isomerase: verification of a ...
|
|
13269
|
Catalytic mechanism of type 2 isopentenyl diphosphate:dimethylallyl diphosphate isomerase: verification of a ...
|
|
13270
|
The vanadate-sensitive ATPase of Streptococcus faecalis pumps potassium in a reconstituted system
|
|
13271
|
The vanadate-sensitive ATPase of Streptococcus faecalis pumps potassium in a reconstituted system
|
|
13272
|
Isolation and characterisation of the pyruvate dehydrogenase complex of anaerobically grown Enterococcus ...
|
|
13273
|
Isolation and characterisation of the pyruvate dehydrogenase complex of anaerobically grown Enterococcus ...
|
|
13274
|
Isolation and characterisation of the pyruvate dehydrogenase complex of anaerobically grown Enterococcus ...
|
|
13275
|
Isolation and characterisation of the pyruvate dehydrogenase complex of anaerobically grown Enterococcus ...
|
|
13276
|
Structural and catalytic characteristics of Escherichia coli adenylate kinase
|
|
13277
|
Structural and catalytic characteristics of Escherichia coli adenylate kinase
|
|
13278
|
Structural and catalytic characteristics of Escherichia coli adenylate kinase
|
|
13279
|
Structural and catalytic characteristics of Escherichia coli adenylate kinase
|
|
13280
|
Structural and catalytic characteristics of Escherichia coli adenylate kinase
|
|
13281
|
Structural and catalytic characteristics of Escherichia coli adenylate kinase
|
|
13282
|
Structural and catalytic characteristics of Escherichia coli adenylate kinase
|
|
13283
|
Structural and catalytic characteristics of Escherichia coli adenylate kinase
|
|
13284
|
Structural and catalytic characteristics of Escherichia coli adenylate kinase
|
|
13285
|
Structural and catalytic characteristics of Escherichia coli adenylate kinase
|
|
13286
|
Phosphofructokinase from Streptococcus lactis
|
|
13287
|
Phosphofructokinase from Streptococcus lactis
|
|
13288
|
Phosphofructokinase from Streptococcus lactis
|
|
13289
|
Phosphofructokinase from Streptococcus lactis
|
|
13290
|
Phosphofructokinase from Streptococcus lactis
|
|
13291
|
Pyruvate kinase from Streptococcus lactis
|
|
13292
|
Pyruvate kinase from Streptococcus lactis
|
|
13293
|
Pyruvate kinase from Streptococcus lactis
|
|
13294
|
Pyruvate kinase from Streptococcus lactis
|
|
13295
|
Pyruvate kinase from Streptococcus lactis
|
|
13296
|
Pyruvate kinase from Streptococcus lactis
|
|
13297
|
Pyruvate kinase from Streptococcus lactis
|
|
13298
|
Pyruvate kinase from Streptococcus lactis
|
|
13299
|
Pyruvate kinase from Streptococcus lactis
|
|
13300
|
Pyruvate kinase from Streptococcus lactis
|
|
13301
|
Pyruvate kinase from Streptococcus lactis
|
|
13302
|
Pyruvate kinase from Streptococcus lactis
|
|
13303
|
Purification and properties of pyruvate kinase from Streptococcus lactis
|
|
13304
|
Purification and properties of pyruvate kinase from Streptococcus lactis
|
|
13305
|
Purification and properties of pyruvate kinase from Streptococcus lactis
|
|
13306
|
Purification and properties of pyruvate kinase from Streptococcus lactis
|
|
13307
|
Purification and properties of pyruvate kinase from Streptococcus lactis
|
|
13308
|
Purification and properties of pyruvate kinase from Streptococcus lactis
|
|
13309
|
Purification and properties of pyruvate kinase from Streptococcus lactis
|
|
13310
|
Purification and properties of pyruvate kinase from Streptococcus lactis
|
|
13311
|
Purification and properties of pyruvate kinase from Streptococcus lactis
|
|
13312
|
Purification and properties of pyruvate kinase from Streptococcus lactis
|
|
13313
|
Purification and properties of pyruvate kinase from Streptococcus lactis
|
|
13314
|
Purification and properties of pyruvate kinase from Streptococcus lactis
|
|
13315
|
Purification and properties of pyruvate kinase from Streptococcus lactis
|
|
13316
|
Purification and properties of pyruvate kinase from Streptococcus lactis
|
|
13317
|
Purification and properties of pyruvate kinase from Streptococcus lactis
|
|
13318
|
Purification and properties of pyruvate kinase from Streptococcus lactis
|
|
13319
|
Purification and properties of pyruvate kinase from Streptococcus lactis
|
|
13320
|
Purification and properties of pyruvate kinase from Streptococcus lactis
|
|
13321
|
Purification and properties of pyruvate kinase from Streptococcus lactis
|
|
13322
|
Purification and properties of pyruvate kinase from Streptococcus lactis
|
|
13323
|
Purification and properties of pyruvate kinase from Streptococcus lactis
|
|
13324
|
Functional Analysis of Conserved Cysteines in Heparan Sulfate N-Deacetylase-N-sulfotransferases
|
|
13325
|
Functional Analysis of Conserved Cysteines in Heparan Sulfate N-Deacetylase-N-sulfotransferases
|
|
13326
|
Functional Analysis of Conserved Cysteines in Heparan Sulfate N-Deacetylase-N-sulfotransferases
|
|
13327
|
Functional Analysis of Conserved Cysteines in Heparan Sulfate N-Deacetylase-N-sulfotransferases
|
|
13328
|
Functional Analysis of Conserved Cysteines in Heparan Sulfate N-Deacetylase-N-sulfotransferases
|
|
13329
|
Functional Analysis of Conserved Cysteines in Heparan Sulfate N-Deacetylase-N-sulfotransferases
|
|
13330
|
Functional Analysis of Conserved Cysteines in Heparan Sulfate N-Deacetylase-N-sulfotransferases
|
|
13331
|
Functional Analysis of Conserved Cysteines in Heparan Sulfate N-Deacetylase-N-sulfotransferases
|
|
13332
|
Functional Analysis of Conserved Cysteines in Heparan Sulfate N-Deacetylase-N-sulfotransferases
|
|
13333
|
Functional Analysis of Conserved Cysteines in Heparan Sulfate N-Deacetylase-N-sulfotransferases
|
|
13334
|
Functional Analysis of Conserved Cysteines in Heparan Sulfate N-Deacetylase-N-sulfotransferases
|
|
13335
|
Functional Analysis of Conserved Cysteines in Heparan Sulfate N-Deacetylase-N-sulfotransferases
|
|
13336
|
Functional Analysis of Conserved Cysteines in Heparan Sulfate N-Deacetylase-N-sulfotransferases
|
|
13337
|
Functional Analysis of Conserved Cysteines in Heparan Sulfate N-Deacetylase-N-sulfotransferases
|
|
13338
|
Functional Analysis of Conserved Cysteines in Heparan Sulfate N-Deacetylase-N-sulfotransferases
|
|
13339
|
Functional Analysis of Conserved Cysteines in Heparan Sulfate N-Deacetylase-N-sulfotransferases
|
|
13340
|
Functional Analysis of Conserved Cysteines in Heparan Sulfate N-Deacetylase-N-sulfotransferases
|
|
13341
|
Functional Analysis of Conserved Cysteines in Heparan Sulfate N-Deacetylase-N-sulfotransferases
|
|
13342
|
Functional Analysis of Conserved Cysteines in Heparan Sulfate N-Deacetylase-N-sulfotransferases
|
|
13343
|
Cysteinyl-tRNA Synthetase from Astragalus Species
|
|
13344
|
Cysteinyl-tRNA Synthetase from Astragalus Species
|
|
13345
|
Cysteinyl-tRNA Synthetase from Astragalus Species
|
|
13346
|
Cysteinyl-tRNA Synthetase from Astragalus Species
|
|
13347
|
Cysteinyl-tRNA Synthetase from Astragalus Species
|
|
13348
|
Cysteinyl-tRNA Synthetase from Astragalus Species
|
|
13349
|
Cysteinyl-tRNA Synthetase from Astragalus Species
|
|
13350
|
Cysteinyl-tRNA Synthetase from Astragalus Species
|
|
13351
|
Cysteinyl-tRNA Synthetase from Astragalus Species
|
|
13352
|
Cysteinyl-tRNA Synthetase from Astragalus Species
|
|
13353
|
Cysteinyl-tRNA Synthetase from Astragalus Species
|
|
13354
|
Cysteinyl-tRNA Synthetase from Phaseolus aureus: Purification and Properties
|
|
13355
|
Cysteinyl-tRNA Synthetase from Phaseolus aureus: Purification and Properties
|
|
13356
|
Cysteinyl-tRNA Synthetase from Phaseolus aureus: Purification and Properties
|
|
13357
|
Cysteinyl-tRNA Synthetase from Phaseolus aureus: Purification and Properties
|
|
13358
|
Cysteinyl-tRNA Synthetase from Phaseolus aureus: Purification and Properties
|
|
13359
|
Cysteinyl-tRNA Synthetase from Phaseolus aureus: Purification and Properties
|
|
13360
|
Cysteinyl-tRNA Synthetase from Phaseolus aureus: Purification and Properties
|
|
13361
|
Cysteinyl-tRNA Synthetase from Phaseolus aureus: Purification and Properties
|
|
13362
|
Cysteinyl-tRNA Synthetase from Phaseolus aureus: Purification and Properties
|
|
13363
|
Inhibition of thymidine kinase by P1-(adenosine-5')-P5-(thymidine-5')-pentaphosphate
|
|
13364
|
Inhibition of thymidine kinase by P1-(adenosine-5')-P5-(thymidine-5')-pentaphosphate
|
|
13365
|
Inhibition of thymidine kinase by P1-(adenosine-5')-P5-(thymidine-5')-pentaphosphate
|
|
13366
|
Inhibition of thymidine kinase by P1-(adenosine-5')-P5-(thymidine-5')-pentaphosphate
|
|
13367
|
Inhibition of thymidine kinase by P1-(adenosine-5')-P5-(thymidine-5')-pentaphosphate
|
|
13368
|
Inhibition of thymidine kinase by P1-(adenosine-5')-P5-(thymidine-5')-pentaphosphate
|
|
13369
|
Inhibition of thymidine kinase by P1-(adenosine-5')-P5-(thymidine-5')-pentaphosphate
|
|
13370
|
Inhibition of thymidine kinase by P1-(adenosine-5')-P5-(thymidine-5')-pentaphosphate
|
|
13371
|
Inhibition of thymidine kinase by P1-(adenosine-5')-P5-(thymidine-5')-pentaphosphate
|
|
13372
|
Inhibition of thymidine kinase by P1-(adenosine-5')-P5-(thymidine-5')-pentaphosphate
|
|
13373
|
Inhibition of thymidine kinase by P1-(adenosine-5')-P5-(thymidine-5')-pentaphosphate
|
|
13374
|
Inhibition of thymidine kinase by P1-(adenosine-5')-P5-(thymidine-5')-pentaphosphate
|
|
13375
|
Inhibition of thymidine kinase by P1-(adenosine-5')-P5-(thymidine-5')-pentaphosphate
|
|
13376
|
Inhibition of thymidine kinase by P1-(adenosine-5')-P5-(thymidine-5')-pentaphosphate
|
|
13377
|
Processing, stability, and kinetic parameters of C5a peptidase from Streptococcus pyogenes
|
|
13378
|
Processing, stability, and kinetic parameters of C5a peptidase from Streptococcus pyogenes
|
|
13379
|
Processing, stability, and kinetic parameters of C5a peptidase from Streptococcus pyogenes
|
|
13380
|
Substitution of cysteine for selenocysteine in the catalytic center of type III iodothyronine deiodinase ...
|
|
13381
|
Substitution of cysteine for selenocysteine in the catalytic center of type III iodothyronine deiodinase ...
|
|
13382
|
Substitution of cysteine for selenocysteine in the catalytic center of type III iodothyronine deiodinase ...
|
|
13383
|
Substitution of cysteine for selenocysteine in the catalytic center of type III iodothyronine deiodinase ...
|
|
13384
|
Substitution of cysteine for selenocysteine in the catalytic center of type III iodothyronine deiodinase ...
|
|
13385
|
Substitution of cysteine for selenocysteine in the catalytic center of type III iodothyronine deiodinase ...
|
|
13386
|
Substitution of cysteine for selenocysteine in the catalytic center of type III iodothyronine deiodinase ...
|
|
13387
|
Substitution of cysteine for selenocysteine in the catalytic center of type III iodothyronine deiodinase ...
|
|
13388
|
Substitution of cysteine for selenocysteine in the catalytic center of type III iodothyronine deiodinase ...
|
|
13389
|
Substitution of cysteine for selenocysteine in the catalytic center of type III iodothyronine deiodinase ...
|
|
13390
|
Substitution of cysteine for selenocysteine in the catalytic center of type III iodothyronine deiodinase ...
|
|
13391
|
Substitution of cysteine for selenocysteine in the catalytic center of type III iodothyronine deiodinase ...
|
|
13392
|
Substitution of cysteine for selenocysteine in the catalytic center of type III iodothyronine deiodinase ...
|
|
13393
|
Substitution of cysteine for selenocysteine in the catalytic center of type III iodothyronine deiodinase ...
|
|
13394
|
Substitution of cysteine for selenocysteine in the catalytic center of type III iodothyronine deiodinase ...
|
|
13395
|
Substitution of cysteine for selenocysteine in the catalytic center of type III iodothyronine deiodinase ...
|
|
13396
|
Substitution of cysteine for selenocysteine in the catalytic center of type III iodothyronine deiodinase ...
|
|
13397
|
Substitution of cysteine for selenocysteine in the catalytic center of type III iodothyronine deiodinase ...
|
|
13398
|
Substitution of cysteine for selenocysteine in the catalytic center of type III iodothyronine deiodinase ...
|
|
13399
|
Substitution of cysteine for selenocysteine in the catalytic center of type III iodothyronine deiodinase ...
|
|
13400
|
Substitution of cysteine for selenocysteine in the catalytic center of type III iodothyronine deiodinase ...
|
|
13401
|
Substitution of cysteine for selenocysteine in the catalytic center of type III iodothyronine deiodinase ...
|
|
13402
|
Substitution of cysteine for selenocysteine in the catalytic center of type III iodothyronine deiodinase ...
|
|
13403
|
Mechanism of action of MK-401 against Fasciola hepatica: inhibition of phosphoglycerate kinase
|
|
13404
|
Mechanism of action of MK-401 against Fasciola hepatica: inhibition of phosphoglycerate kinase
|
|
13405
|
Mechanism of action of MK-401 against Fasciola hepatica: inhibition of phosphoglycerate kinase
|
|
13406
|
Mechanism of action of MK-401 against Fasciola hepatica: inhibition of phosphoglycerate kinase
|
|
13407
|
Mechanism of action of MK-401 against Fasciola hepatica: inhibition of phosphoglycerate kinase
|
|
13408
|
Steady-state kinetics of the hypoxanthine phosphoribosyltransferase from Trypanosoma cruzi
|
|
13409
|
Steady-state kinetics of the hypoxanthine phosphoribosyltransferase from Trypanosoma cruzi
|
|
13410
|
Steady-state kinetics of the hypoxanthine phosphoribosyltransferase from Trypanosoma cruzi
|
|
13411
|
Steady-state kinetics of the hypoxanthine phosphoribosyltransferase from Trypanosoma cruzi
|
|
13412
|
Steady-state kinetics of the hypoxanthine phosphoribosyltransferase from Trypanosoma cruzi
|
|
13413
|
Steady-state kinetics of the hypoxanthine phosphoribosyltransferase from Trypanosoma cruzi
|
|
13414
|
Steady-state kinetics of the hypoxanthine phosphoribosyltransferase from Trypanosoma cruzi
|
|
13415
|
Steady-state kinetics of the hypoxanthine phosphoribosyltransferase from Trypanosoma cruzi
|
|
13416
|
Steady-state kinetics of the hypoxanthine phosphoribosyltransferase from Trypanosoma cruzi
|
|
13417
|
Steady-state kinetics of the hypoxanthine phosphoribosyltransferase from Trypanosoma cruzi
|
|
13418
|
Steady-state kinetics of the hypoxanthine phosphoribosyltransferase from Trypanosoma cruzi
|
|
13419
|
Steady-state kinetics of the hypoxanthine phosphoribosyltransferase from Trypanosoma cruzi
|
|
13420
|
Steady-state kinetics of the hypoxanthine phosphoribosyltransferase from Trypanosoma cruzi
|
|
13421
|
Steady-state kinetics of the hypoxanthine phosphoribosyltransferase from Trypanosoma cruzi
|
|
13422
|
Steady-state kinetics of the hypoxanthine phosphoribosyltransferase from Trypanosoma cruzi
|
|
13423
|
Steady-state kinetics of the hypoxanthine phosphoribosyltransferase from Trypanosoma cruzi
|
|
13424
|
Steady-state kinetics of the hypoxanthine phosphoribosyltransferase from Trypanosoma cruzi
|
|
13425
|
Steady-state kinetics of the hypoxanthine phosphoribosyltransferase from Trypanosoma cruzi
|
|
13426
|
Steady-state kinetics of the hypoxanthine phosphoribosyltransferase from Trypanosoma cruzi
|
|
13427
|
Steady-state kinetics of the hypoxanthine phosphoribosyltransferase from Trypanosoma cruzi
|
|
13428
|
Characterization of transsulfuration and cysteine biosynthetic pathways in the protozoan hemoflagellate, ...
|
|
13429
|
Characterization of transsulfuration and cysteine biosynthetic pathways in the protozoan hemoflagellate, ...
|
|
13430
|
Characterization of transsulfuration and cysteine biosynthetic pathways in the protozoan hemoflagellate, ...
|
|
13431
|
Characterization of transsulfuration and cysteine biosynthetic pathways in the protozoan hemoflagellate, ...
|
|
13432
|
Characterization of transsulfuration and cysteine biosynthetic pathways in the protozoan hemoflagellate, ...
|
|
13433
|
Characterization of transsulfuration and cysteine biosynthetic pathways in the protozoan hemoflagellate, ...
|
|
13434
|
Characterization of transsulfuration and cysteine biosynthetic pathways in the protozoan hemoflagellate, ...
|
|
13435
|
Characterization of transsulfuration and cysteine biosynthetic pathways in the protozoan hemoflagellate, ...
|
|
13436
|
Negative cooperativity in pyridoxal 5`-phosphate binding and enzyme activity of 6-phosphoglucose isomerase ...
|
|
13437
|
Negative cooperativity in pyridoxal 5`-phosphate binding and enzyme activity of 6-phosphoglucose isomerase ...
|
|
13438
|
Negative cooperativity in pyridoxal 5`-phosphate binding and enzyme activity of 6-phosphoglucose isomerase ...
|
|
13439
|
Negative cooperativity in pyridoxal 5`-phosphate binding and enzyme activity of 6-phosphoglucose isomerase ...
|
|
13440
|
Negative cooperativity in pyridoxal 5`-phosphate binding and enzyme activity of 6-phosphoglucose isomerase ...
|
|
13441
|
Negative cooperativity in pyridoxal 5`-phosphate binding and enzyme activity of 6-phosphoglucose isomerase ...
|
|
13442
|
Negative cooperativity in pyridoxal 5`-phosphate binding and enzyme activity of 6-phosphoglucose isomerase ...
|
|
13443
|
Glucose-6-phosphate isomerase from Ehrlich ascites tumor cells: purification and some properties.
|
|
13444
|
Glucose-6-phosphate isomerase from Ehrlich ascites tumor cells: purification and some properties.
|
|
13445
|
Mechanism of action of enolase: effect of the beta-hydroxy group on the rate of dissociation of the ...
|
|
13446
|
Mechanism of action of enolase: effect of the beta-hydroxy group on the rate of dissociation of the ...
|
|
13447
|
Mechanism of action of enolase: effect of the beta-hydroxy group on the rate of dissociation of the ...
|
|
13448
|
Mechanism of action of enolase: effect of the beta-hydroxy group on the rate of dissociation of the ...
|
|
13449
|
Mechanism of action of enolase: effect of the beta-hydroxy group on the rate of dissociation of the ...
|
|
13450
|
Mechanism of action of enolase: effect of the beta-hydroxy group on the rate of dissociation of the ...
|
|
13451
|
Mechanism of action of enolase: effect of the beta-hydroxy group on the rate of dissociation of the ...
|
|
13452
|
Mechanism of action of enolase: effect of the beta-hydroxy group on the rate of dissociation of the ...
|
|
13453
|
Mechanism of action of enolase: effect of the beta-hydroxy group on the rate of dissociation of the ...
|
|
13454
|
Mechanism of action of enolase: effect of the beta-hydroxy group on the rate of dissociation of the ...
|
|
13455
|
Mechanism of action of enolase: effect of the beta-hydroxy group on the rate of dissociation of the ...
|
|
13456
|
Mechanism of action of enolase: effect of the beta-hydroxy group on the rate of dissociation of the ...
|
|
13457
|
On the specificity of phospho-N-acetylmuramyl-pentapeptide translocase. The peptide subunit of uridine ...
|
|
13458
|
On the specificity of phospho-N-acetylmuramyl-pentapeptide translocase. The peptide subunit of uridine ...
|
|
13459
|
On the specificity of phospho-N-acetylmuramyl-pentapeptide translocase. The peptide subunit of uridine ...
|
|
13460
|
On the specificity of phospho-N-acetylmuramyl-pentapeptide translocase. The peptide subunit of uridine ...
|
|
13461
|
On the specificity of phospho-N-acetylmuramyl-pentapeptide translocase. The peptide subunit of uridine ...
|
|
13462
|
On the specificity of phospho-N-acetylmuramyl-pentapeptide translocase. The peptide subunit of uridine ...
|
|
13463
|
On the specificity of phospho-N-acetylmuramyl-pentapeptide translocase. The peptide subunit of uridine ...
|
|
13464
|
On the specificity of phospho-N-acetylmuramyl-pentapeptide translocase. The peptide subunit of uridine ...
|
|
13465
|
Reactive amino acid residues involved in glutamate-binding of human glutamate dehydrogenase isozymes
|
|
13466
|
Reactive amino acid residues involved in glutamate-binding of human glutamate dehydrogenase isozymes
|
|
13467
|
Reactive amino acid residues involved in glutamate-binding of human glutamate dehydrogenase isozymes
|
|
13468
|
Reactive amino acid residues involved in glutamate-binding of human glutamate dehydrogenase isozymes
|
|
13469
|
Reactive amino acid residues involved in glutamate-binding of human glutamate dehydrogenase isozymes
|
|
13470
|
Reactive amino acid residues involved in glutamate-binding of human glutamate dehydrogenase isozymes
|
|
13471
|
Reactive amino acid residues involved in glutamate-binding of human glutamate dehydrogenase isozymes
|
|
13472
|
Reactive amino acid residues involved in glutamate-binding of human glutamate dehydrogenase isozymes
|
|
13473
|
Reactive amino acid residues involved in glutamate-binding of human glutamate dehydrogenase isozymes
|
|
13474
|
Reactive amino acid residues involved in glutamate-binding of human glutamate dehydrogenase isozymes
|
|
13475
|
Reactive amino acid residues involved in glutamate-binding of human glutamate dehydrogenase isozymes
|
|
13476
|
Reactive amino acid residues involved in glutamate-binding of human glutamate dehydrogenase isozymes
|
|
13477
|
Reactive amino acid residues involved in glutamate-binding of human glutamate dehydrogenase isozymes
|
|
13478
|
Reactive amino acid residues involved in glutamate-binding of human glutamate dehydrogenase isozymes
|
|
13479
|
Reactive amino acid residues involved in glutamate-binding of human glutamate dehydrogenase isozymes
|
|
13480
|
Reactive amino acid residues involved in glutamate-binding of human glutamate dehydrogenase isozymes
|
|
13481
|
Reactive amino acid residues involved in glutamate-binding of human glutamate dehydrogenase isozymes
|
|
13482
|
Reactive amino acid residues involved in glutamate-binding of human glutamate dehydrogenase isozymes
|
|
13483
|
Reactive amino acid residues involved in glutamate-binding of human glutamate dehydrogenase isozymes
|
|
13484
|
Reactive amino acid residues involved in glutamate-binding of human glutamate dehydrogenase isozymes
|
|
13485
|
Reactive amino acid residues involved in glutamate-binding of human glutamate dehydrogenase isozymes
|
|
13486
|
Reactive amino acid residues involved in glutamate-binding of human glutamate dehydrogenase isozymes
|
|
13487
|
Reactive amino acid residues involved in glutamate-binding of human glutamate dehydrogenase isozymes
|
|
13488
|
Reactive amino acid residues involved in glutamate-binding of human glutamate dehydrogenase isozymes
|
|
13489
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13490
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13491
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13492
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13493
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13494
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13495
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13496
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13497
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13498
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13499
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13500
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13501
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13502
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13503
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13504
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13505
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13506
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13507
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13508
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13509
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13510
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13511
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13512
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13513
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13514
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13515
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13516
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13517
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13518
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13519
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13520
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13521
|
A rapid screening assay for inhibitors of 11beta-hydroxysteroid dehydrogenases (11beta-HSD): flavanone ...
|
|
13522
|
Kinetic properties of NAD-dependent glyceraldehyde-3-phosphate dehydrogenase from the host fraction of soybean ...
|
|
13523
|
Kinetic properties of NAD-dependent glyceraldehyde-3-phosphate dehydrogenase from the host fraction of soybean ...
|
|
13524
|
Kinetic properties of NAD-dependent glyceraldehyde-3-phosphate dehydrogenase from the host fraction of soybean ...
|
|
13525
|
Kinetic properties of NAD-dependent glyceraldehyde-3-phosphate dehydrogenase from the host fraction of soybean ...
|
|
13526
|
Kinetic properties of NAD-dependent glyceraldehyde-3-phosphate dehydrogenase from the host fraction of soybean ...
|
|
13527
|
Kinetic properties of NAD-dependent glyceraldehyde-3-phosphate dehydrogenase from the host fraction of soybean ...
|
|
13528
|
Kinetic properties of NAD-dependent glyceraldehyde-3-phosphate dehydrogenase from the host fraction of soybean ...
|
|
13529
|
Kinetic properties of NAD-dependent glyceraldehyde-3-phosphate dehydrogenase from the host fraction of soybean ...
|
|
13530
|
Kinetic properties of NAD-dependent glyceraldehyde-3-phosphate dehydrogenase from the host fraction of soybean ...
|
|
13531
|
Kinetic properties of NAD-dependent glyceraldehyde-3-phosphate dehydrogenase from the host fraction of soybean ...
|
|
13532
|
Kinetic properties of NAD-dependent glyceraldehyde-3-phosphate dehydrogenase from the host fraction of soybean ...
|
|
13533
|
Kinetic properties of NAD-dependent glyceraldehyde-3-phosphate dehydrogenase from the host fraction of soybean ...
|
|
13534
|
Kinetic properties of NAD-dependent glyceraldehyde-3-phosphate dehydrogenase from the host fraction of soybean ...
|
|
13535
|
Kinetic properties of NAD-dependent glyceraldehyde-3-phosphate dehydrogenase from the host fraction of soybean ...
|
|
13536
|
Purification and properties of three NAD(P)+ isozymes of L-glutamate dehydrogenase of Chlamydomonas ...
|
|
13537
|
Purification and properties of three NAD(P)+ isozymes of L-glutamate dehydrogenase of Chlamydomonas ...
|
|
13538
|
Purification and properties of three NAD(P)+ isozymes of L-glutamate dehydrogenase of Chlamydomonas ...
|
|
13539
|
Purification and properties of three NAD(P)+ isozymes of L-glutamate dehydrogenase of Chlamydomonas ...
|
|
13540
|
Purification and properties of three NAD(P)+ isozymes of L-glutamate dehydrogenase of Chlamydomonas ...
|
|
13541
|
Purification and properties of three NAD(P)+ isozymes of L-glutamate dehydrogenase of Chlamydomonas ...
|
|
13542
|
Purification and properties of three NAD(P)+ isozymes of L-glutamate dehydrogenase of Chlamydomonas ...
|
|
13543
|
Purification and properties of three NAD(P)+ isozymes of L-glutamate dehydrogenase of Chlamydomonas ...
|
|
13544
|
Purification and properties of three NAD(P)+ isozymes of L-glutamate dehydrogenase of Chlamydomonas ...
|
|
13545
|
Purification and properties of three NAD(P)+ isozymes of L-glutamate dehydrogenase of Chlamydomonas ...
|
|
13546
|
Purification and properties of three NAD(P)+ isozymes of L-glutamate dehydrogenase of Chlamydomonas ...
|
|
13547
|
Purification and properties of three NAD(P)+ isozymes of L-glutamate dehydrogenase of Chlamydomonas ...
|
|
13548
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13549
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13550
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13551
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13552
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13553
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13554
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13555
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13556
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13557
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13558
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13559
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13560
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13561
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13562
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13563
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13564
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13565
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13566
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13567
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13568
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13569
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13570
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13571
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13572
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13573
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13574
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13575
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13576
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13577
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13578
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13579
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13580
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13581
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13582
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13583
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13584
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13585
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13586
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13587
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13588
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13589
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13590
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13591
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13592
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13593
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13594
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13595
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13596
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13597
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13598
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13599
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13600
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13601
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13602
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13603
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13604
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13605
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13606
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13607
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13608
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13609
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13610
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13611
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13612
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13613
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13614
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13615
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13616
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13617
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13618
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13619
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13620
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13621
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13622
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13623
|
Laboratory-evolved vanillyl-alcohol oxidase produces natural vanillin
|
|
13624
|
Ornithine carbamoyltransferase from the extreme thermophile Thermus thermophilus--analysis of the gene and ...
|
|
13625
|
Ornithine carbamoyltransferase from the extreme thermophile Thermus thermophilus--analysis of the gene and ...
|
|
13626
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13627
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13628
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13629
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13630
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13631
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13632
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13633
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13634
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13635
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13636
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13637
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13638
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13639
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13640
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13641
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13642
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13643
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13644
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13645
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13646
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13647
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13648
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13649
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13650
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13651
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13652
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13653
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13654
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13655
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13656
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13657
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13658
|
Characterization of four beta-lactamases produced by Staphylococcus aureus
|
|
13659
|
Identification of genus Nitrosovibrio, ammonia-oxidizing bacteria, by comparison of N-terminal amino acid ...
|
|
13660
|
Identification of genus Nitrosovibrio, ammonia-oxidizing bacteria, by comparison of N-terminal amino acid ...
|
|
13661
|
Identification of genus Nitrosovibrio, ammonia-oxidizing bacteria, by comparison of N-terminal amino acid ...
|
|
13662
|
Identification of genus Nitrosovibrio, ammonia-oxidizing bacteria, by comparison of N-terminal amino acid ...
|
|
13663
|
Identification of genus Nitrosovibrio, ammonia-oxidizing bacteria, by comparison of N-terminal amino acid ...
|
|
13664
|
Identification of genus Nitrosovibrio, ammonia-oxidizing bacteria, by comparison of N-terminal amino acid ...
|
|
13665
|
Identification of genus Nitrosovibrio, ammonia-oxidizing bacteria, by comparison of N-terminal amino acid ...
|
|
13666
|
Identification of genus Nitrosovibrio, ammonia-oxidizing bacteria, by comparison of N-terminal amino acid ...
|
|
13667
|
Identification of genus Nitrosovibrio, ammonia-oxidizing bacteria, by comparison of N-terminal amino acid ...
|
|
13668
|
Identification of genus Nitrosovibrio, ammonia-oxidizing bacteria, by comparison of N-terminal amino acid ...
|
|
13669
|
Identification of genus Nitrosovibrio, ammonia-oxidizing bacteria, by comparison of N-terminal amino acid ...
|
|
13670
|
Kinetic and chemical mechanism of alpha-isopropylmalate synthase from Mycobacterium tuberculosis
|
|
13671
|
Kinetic and chemical mechanism of alpha-isopropylmalate synthase from Mycobacterium tuberculosis
|
|
13672
|
Kinetic and chemical mechanism of alpha-isopropylmalate synthase from Mycobacterium tuberculosis
|
|
13673
|
Kinetic and chemical mechanism of alpha-isopropylmalate synthase from Mycobacterium tuberculosis
|
|
13674
|
Kinetic and chemical mechanism of alpha-isopropylmalate synthase from Mycobacterium tuberculosis
|
|
13675
|
Kinetic and chemical mechanism of alpha-isopropylmalate synthase from Mycobacterium tuberculosis
|
|
13676
|
Kinetic and chemical mechanism of alpha-isopropylmalate synthase from Mycobacterium tuberculosis
|
|
13677
|
Kinetic and chemical mechanism of alpha-isopropylmalate synthase from Mycobacterium tuberculosis
|
|
13678
|
Kinetic and chemical mechanism of alpha-isopropylmalate synthase from Mycobacterium tuberculosis
|
|
13679
|
Kinetic and chemical mechanism of alpha-isopropylmalate synthase from Mycobacterium tuberculosis
|
|
13680
|
Kinetic and chemical mechanism of alpha-isopropylmalate synthase from Mycobacterium tuberculosis
|
|
13681
|
Kinetic and chemical mechanism of alpha-isopropylmalate synthase from Mycobacterium tuberculosis
|
|
13682
|
Kinetic and chemical mechanism of alpha-isopropylmalate synthase from Mycobacterium tuberculosis
|
|
13683
|
Kinetic and chemical mechanism of alpha-isopropylmalate synthase from Mycobacterium tuberculosis
|
|
13684
|
Kinetic and chemical mechanism of alpha-isopropylmalate synthase from Mycobacterium tuberculosis
|
|
13685
|
Kinetic and chemical mechanism of alpha-isopropylmalate synthase from Mycobacterium tuberculosis
|
|
13686
|
Kinetic and chemical mechanism of alpha-isopropylmalate synthase from Mycobacterium tuberculosis
|
|
13687
|
Kinetic and chemical mechanism of alpha-isopropylmalate synthase from Mycobacterium tuberculosis
|
|
13688
|
Kinetic and chemical mechanism of alpha-isopropylmalate synthase from Mycobacterium tuberculosis
|
|
13689
|
Kinetic and chemical mechanism of alpha-isopropylmalate synthase from Mycobacterium tuberculosis
|
|
13690
|
Characterization of the enolase isozymes of rabbit brain: kinetic differences between mammalian and yeast ...
|
|
13691
|
Characterization of the enolase isozymes of rabbit brain: kinetic differences between mammalian and yeast ...
|
|
13692
|
Characterization of the enolase isozymes of rabbit brain: kinetic differences between mammalian and yeast ...
|
|
13693
|
Characterization of the enolase isozymes of rabbit brain: kinetic differences between mammalian and yeast ...
|
|
13694
|
Effects of fructose 1,6-bisphosphate on the activation of yeast phosphofructokinase by fructose ...
|
|
13695
|
Effects of fructose 1,6-bisphosphate on the activation of yeast phosphofructokinase by fructose ...
|
|
13696
|
Effects of fructose 1,6-bisphosphate on the activation of yeast phosphofructokinase by fructose ...
|
|
13697
|
Effects of fructose 1,6-bisphosphate on the activation of yeast phosphofructokinase by fructose ...
|
|
13698
|
Inactivation kinetics of mushroom tyrosinase by cetylpyridinium chloride
|
|
13699
|
Human peroxisomal multifunctional enzyme type 2. Site-directed mutagenesis studies show the importance of two ...
|
|
13700
|
Human peroxisomal multifunctional enzyme type 2. Site-directed mutagenesis studies show the importance of two ...
|
|
13701
|
Human peroxisomal multifunctional enzyme type 2. Site-directed mutagenesis studies show the importance of two ...
|
|
13702
|
Human peroxisomal multifunctional enzyme type 2. Site-directed mutagenesis studies show the importance of two ...
|
|
13703
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13704
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13705
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13706
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13707
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13708
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13709
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13710
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13711
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13712
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13713
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13714
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13715
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13716
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13717
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13718
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13719
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13720
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13721
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13722
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13723
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13724
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13725
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13726
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13727
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13728
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13729
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13730
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13731
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13732
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13733
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13734
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13735
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13736
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13737
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13738
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13739
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13740
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13741
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13742
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13743
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13744
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13745
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13746
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13747
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13748
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13749
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13750
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13751
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13752
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13753
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13754
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13755
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13756
|
Involvement of a flavosemiquinone in the enzymatic oxidation of nitroalkanes catalyzed by 2-nitropropane ...
|
|
13757
|
Kinetic and Stereochemical Comparison of Wild-type and Active-Site K145Q Mutant Enzyme of Bacterial D-Amino ...
|
|
13758
|
Kinetic and Stereochemical Comparison of Wild-type and Active-Site K145Q Mutant Enzyme of Bacterial D-Amino ...
|
|
13759
|
Kinetic and Stereochemical Comparison of Wild-type and Active-Site K145Q Mutant Enzyme of Bacterial D-Amino ...
|
|
13760
|
Kinetic and Stereochemical Comparison of Wild-type and Active-Site K145Q Mutant Enzyme of Bacterial D-Amino ...
|
|
13761
|
Kinetic and Stereochemical Comparison of Wild-type and Active-Site K145Q Mutant Enzyme of Bacterial D-Amino ...
|
|
13762
|
Kinetic and Stereochemical Comparison of Wild-type and Active-Site K145Q Mutant Enzyme of Bacterial D-Amino ...
|
|
13763
|
Characterization of recombinant human aldolase B and purification by metal chelate chromatography
|
|
13764
|
Characterization of recombinant human aldolase B and purification by metal chelate chromatography
|
|
13765
|
Characterization of recombinant human aldolase B and purification by metal chelate chromatography
|
|
13766
|
Characterization of recombinant human aldolase B and purification by metal chelate chromatography
|
|
13767
|
Characterization of recombinant human aldolase B and purification by metal chelate chromatography
|
|
13768
|
Characterization of recombinant human aldolase B and purification by metal chelate chromatography
|
|
13769
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13770
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13771
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13772
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13773
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13774
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13775
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13776
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13777
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13778
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13779
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13780
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13781
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13782
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13783
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13784
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13785
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13786
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13787
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13788
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13789
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13790
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13791
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13792
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13793
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13794
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13795
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13796
|
Critical role of Lys212 and Tyr140 in porcine NADP-dependent isocitrate dehydrogenase
|
|
13893
|
Studies on the regulation of enolases and compartmentation of cytosolic enzymes in Saccharomyces cerevisiae
|
|
13894
|
Studies on the regulation of enolases and compartmentation of cytosolic enzymes in Saccharomyces cerevisiae
|
|
13895
|
Studies on the regulation of enolases and compartmentation of cytosolic enzymes in Saccharomyces cerevisiae
|
|
13896
|
Studies on the regulation of enolases and compartmentation of cytosolic enzymes in Saccharomyces cerevisiae
|
|
13897
|
Studies on the regulation of enolases and compartmentation of cytosolic enzymes in Saccharomyces cerevisiae
|
|
13898
|
Studies on the regulation of enolases and compartmentation of cytosolic enzymes in Saccharomyces cerevisiae
|
|
13899
|
Characterization of alpha alpha, beta beta, gamma gamma and alpha gamma human enolase isozymes, and ...
|
|
13900
|
Characterization of alpha alpha, beta beta, gamma gamma and alpha gamma human enolase isozymes, and ...
|
|
13901
|
Characterization of alpha alpha, beta beta, gamma gamma and alpha gamma human enolase isozymes, and ...
|
|
13902
|
Characterization of alpha alpha, beta beta, gamma gamma and alpha gamma human enolase isozymes, and ...
|
|
13903
|
Evolution of enzymatic activities in the enolase superfamily: identification of the general acid catalyst in ...
|
|
13904
|
Evolution of enzymatic activities in the enolase superfamily: identification of the general acid catalyst in ...
|
|
13905
|
Evolution of enzymatic activities in the enolase superfamily: identification of the general acid catalyst in ...
|
|
13906
|
Evolution of enzymatic activities in the enolase superfamily: identification of the general acid catalyst in ...
|
|
13907
|
Evolution of enzymatic activities in the enolase superfamily: identification of the general acid catalyst in ...
|
|
13908
|
Evolution of enzymatic activities in the enolase superfamily: identification of the general acid catalyst in ...
|
|
13909
|
Evolution of enzymatic activities in the enolase superfamily: identification of the general acid catalyst in ...
|
|
13910
|
Evolution of enzymatic activities in the enolase superfamily: identification of the general acid catalyst in ...
|
|
13911
|
Evolution of enzymatic activities in the enolase superfamily: identification of the general acid catalyst in ...
|
|
13912
|
Functional and structural changes due to a serine to alanine mutation in the active-site flap of enolase
|
|
13913
|
Functional and structural changes due to a serine to alanine mutation in the active-site flap of enolase
|
|
13914
|
Functional and structural changes due to a serine to alanine mutation in the active-site flap of enolase
|
|
13915
|
Functional and structural changes due to a serine to alanine mutation in the active-site flap of enolase
|
|
13916
|
Functional and structural changes due to a serine to alanine mutation in the active-site flap of enolase
|
|
13917
|
Functional and structural changes due to a serine to alanine mutation in the active-site flap of enolase
|
|
13918
|
Functional and structural changes due to a serine to alanine mutation in the active-site flap of enolase
|
|
13919
|
Functional and structural changes due to a serine to alanine mutation in the active-site flap of enolase
|
|
13920
|
Functional and structural changes due to a serine to alanine mutation in the active-site flap of enolase
|
|
13922
|
Fructose-1,6-diphosphate-dependent lactate dehydrogenase from a cariogenic streptococcus: purification and ...
|
|
13923
|
Fructose-1,6-diphosphate-dependent lactate dehydrogenase from a cariogenic streptococcus: purification and ...
|
|
13924
|
Fructose-1,6-diphosphate-dependent lactate dehydrogenase from a cariogenic streptococcus: purification and ...
|
|
13925
|
Fructose-1,6-diphosphate-dependent lactate dehydrogenase from a cariogenic streptococcus: purification and ...
|
|
13926
|
Rat Guanidinoacetate Methyltransferase- Effect of Site-Directed Alteration of an Aspartic Acid Residue that is ...
|
|
13927
|
Rat Guanidinoacetate Methyltransferase- Effect of Site-Directed Alteration of an Aspartic Acid Residue that is ...
|
|
13928
|
Rat Guanidinoacetate Methyltransferase- Effect of Site-Directed Alteration of an Aspartic Acid Residue that is ...
|
|
13929
|
Rat Guanidinoacetate Methyltransferase- Effect of Site-Directed Alteration of an Aspartic Acid Residue that is ...
|
|
13930
|
Rat Guanidinoacetate Methyltransferase- Effect of Site-Directed Alteration of an Aspartic Acid Residue that is ...
|
|
13931
|
Rat Guanidinoacetate Methyltransferase- Effect of Site-Directed Alteration of an Aspartic Acid Residue that is ...
|
|
13932
|
Rat Guanidinoacetate Methyltransferase- Effect of Site-Directed Alteration of an Aspartic Acid Residue that is ...
|
|
13933
|
Rat Guanidinoacetate Methyltransferase- Effect of Site-Directed Alteration of an Aspartic Acid Residue that is ...
|
|
13934
|
Rat Guanidinoacetate Methyltransferase- Effect of Site-Directed Alteration of an Aspartic Acid Residue that is ...
|
|
13935
|
Rat Guanidinoacetate Methyltransferase- Effect of Site-Directed Alteration of an Aspartic Acid Residue that is ...
|
|
13936
|
Rat Guanidinoacetate Methyltransferase- Effect of Site-Directed Alteration of an Aspartic Acid Residue that is ...
|
|
13937
|
Rat Guanidinoacetate Methyltransferase- Effect of Site-Directed Alteration of an Aspartic Acid Residue that is ...
|
|
13938
|
Rat Guanidinoacetate Methyltransferase- Effect of Site-Directed Alteration of an Aspartic Acid Residue that is ...
|
|
13939
|
Rat Guanidinoacetate Methyltransferase- Effect of Site-Directed Alteration of an Aspartic Acid Residue that is ...
|
|
13940
|
Rat Guanidinoacetate Methyltransferase- Effect of Site-Directed Alteration of an Aspartic Acid Residue that is ...
|
|
13941
|
Rat Guanidinoacetate Methyltransferase- Effect of Site-Directed Alteration of an Aspartic Acid Residue that is ...
|
|
13942
|
Rat Guanidinoacetate Methyltransferase- Effect of Site-Directed Alteration of an Aspartic Acid Residue that is ...
|
|
13943
|
Rat Guanidinoacetate Methyltransferase- Effect of Site-Directed Alteration of an Aspartic Acid Residue that is ...
|
|
13944
|
Rat Guanidinoacetate Methyltransferase- Effect of Site-Directed Alteration of an Aspartic Acid Residue that is ...
|
|
13945
|
Structural determinants of substrate binding to Bacillus cereus metallo-beta-lactamase
|
|
13946
|
Structural determinants of substrate binding to Bacillus cereus metallo-beta-lactamase
|
|
13947
|
Structural determinants of substrate binding to Bacillus cereus metallo-beta-lactamase
|
|
13984
|
Lys-D48 is required for charge stabilization, rapid flavin reduction, and internal electron transfer in the ...
|
|
13985
|
Lys-D48 is required for charge stabilization, rapid flavin reduction, and internal electron transfer in the ...
|
|
13986
|
Lys-D48 is required for charge stabilization, rapid flavin reduction, and internal electron transfer in the ...
|
|
13987
|
Lys-D48 is required for charge stabilization, rapid flavin reduction, and internal electron transfer in the ...
|
|
13988
|
Lys-D48 is required for charge stabilization, rapid flavin reduction, and internal electron transfer in the ...
|
|
13989
|
Lys-D48 is required for charge stabilization, rapid flavin reduction, and internal electron transfer in the ...
|
|
13990
|
Lys-D48 is required for charge stabilization, rapid flavin reduction, and internal electron transfer in the ...
|
|
13991
|
Lys-D48 is required for charge stabilization, rapid flavin reduction, and internal electron transfer in the ...
|
|
13992
|
Lys-D48 is required for charge stabilization, rapid flavin reduction, and internal electron transfer in the ...
|
|
13993
|
Lys-D48 is required for charge stabilization, rapid flavin reduction, and internal electron transfer in the ...
|
|
13994
|
Lys-D48 is required for charge stabilization, rapid flavin reduction, and internal electron transfer in the ...
|
|
13995
|
Lys-D48 is required for charge stabilization, rapid flavin reduction, and internal electron transfer in the ...
|
|
13996
|
Lys-D48 is required for charge stabilization, rapid flavin reduction, and internal electron transfer in the ...
|
|
13997
|
Steady-state kinetic analysis of the quinoprotein methylamine dehydrogenase from Paracoccus denitrificans
|
|
13998
|
Steady-state kinetic analysis of the quinoprotein methylamine dehydrogenase from Paracoccus denitrificans
|
|
13999
|
Steady-state kinetic analysis of the quinoprotein methylamine dehydrogenase from Paracoccus denitrificans
|
|
14000
|
Steady-state kinetic analysis of the quinoprotein methylamine dehydrogenase from Paracoccus denitrificans
|





 If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.
If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.