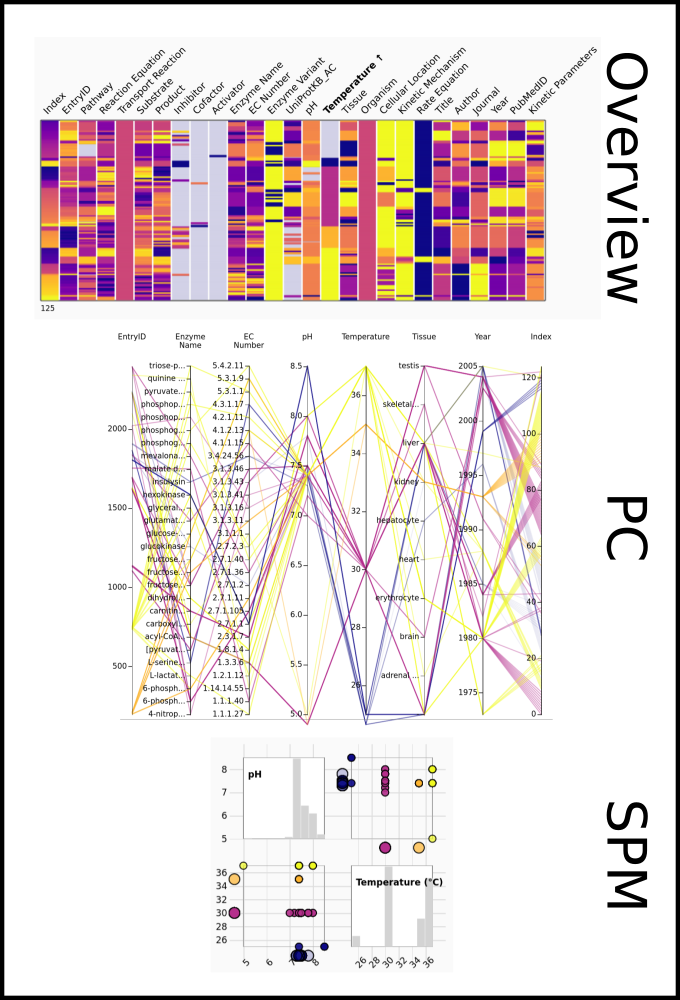
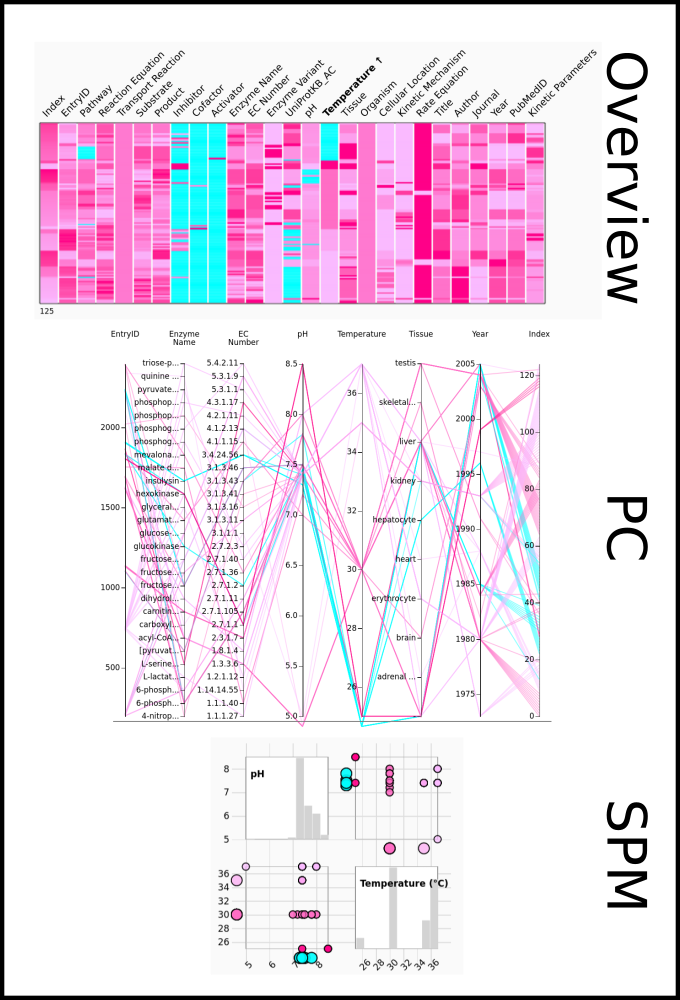
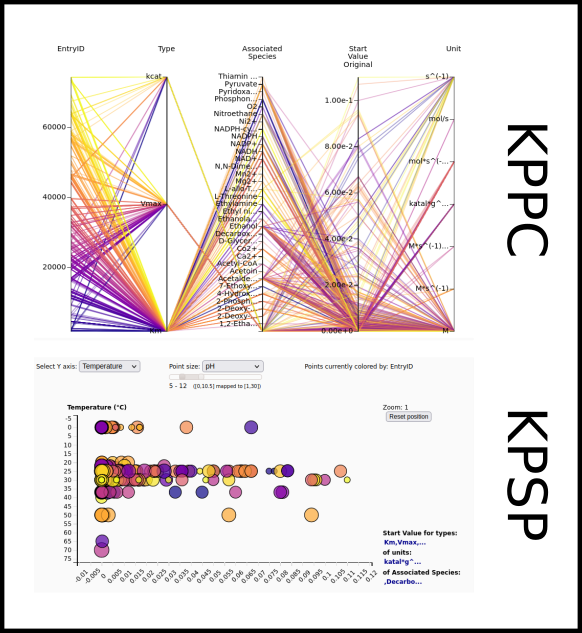
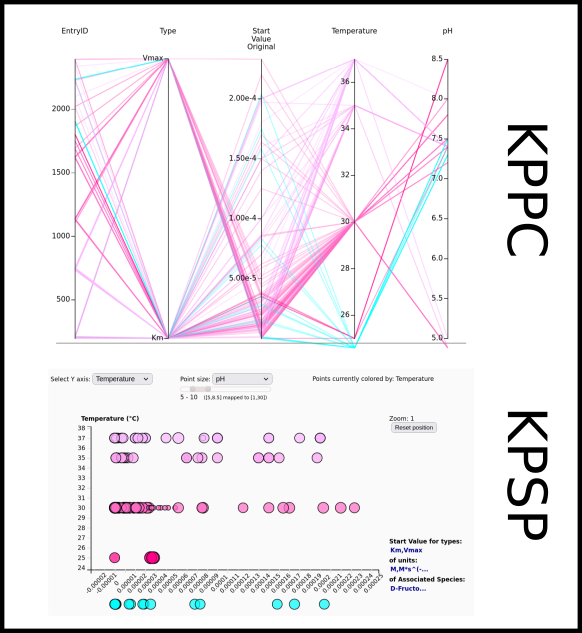
To improve SABIO-RK data interoperability, semantic markup was added to web pages as described and defined by
This structured information makes it easier to discover, collate and analyse our data.
|
72001
|
Characterization of human fructose-1,6-bisphosphatase in control and deficient tissues.
|
|
72002
|
Characterization of human fructose-1,6-bisphosphatase in control and deficient tissues.
|
|
72003
|
Characterization of human fructose-1,6-bisphosphatase in control and deficient tissues.
|
|
72004
|
Characterization of human fructose-1,6-bisphosphatase in control and deficient tissues.
|
|
72005
|
Characterization of human fructose-1,6-bisphosphatase in control and deficient tissues.
|
|
72006
|
Characterization of human fructose-1,6-bisphosphatase in control and deficient tissues.
|
|
72007
|
Sesquiterpene synthases Cop4 and Cop6 from Coprinus cinereus: catalytic promiscuity and cyclization of ...
|
|
72008
|
Sesquiterpene synthases Cop4 and Cop6 from Coprinus cinereus: catalytic promiscuity and cyclization of ...
|
|
72009
|
Sesquiterpene synthases Cop4 and Cop6 from Coprinus cinereus: catalytic promiscuity and cyclization of ...
|
|
72010
|
Sesquiterpene synthases Cop4 and Cop6 from Coprinus cinereus: catalytic promiscuity and cyclization of ...
|
|
72011
|
Sesquiterpene synthases Cop4 and Cop6 from Coprinus cinereus: catalytic promiscuity and cyclization of ...
|
|
72012
|
Sesquiterpene synthases Cop4 and Cop6 from Coprinus cinereus: catalytic promiscuity and cyclization of ...
|
|
72013
|
Sesquiterpene synthases Cop4 and Cop6 from Coprinus cinereus: catalytic promiscuity and cyclization of ...
|
|
72014
|
Truncation of limonene synthase preprotein provides a fully active ""pseudomature"" form of this monoterpene ...
|
|
72015
|
Truncation of limonene synthase preprotein provides a fully active ""pseudomature"" form of this monoterpene ...
|
|
72016
|
Truncation of limonene synthase preprotein provides a fully active ""pseudomature"" form of this monoterpene ...
|
|
72017
|
Truncation of limonene synthase preprotein provides a fully active ""pseudomature"" form of this monoterpene ...
|
|
72018
|
Truncation of limonene synthase preprotein provides a fully active ""pseudomature"" form of this monoterpene ...
|
|
72019
|
Truncation of limonene synthase preprotein provides a fully active ""pseudomature"" form of this monoterpene ...
|
|
72020
|
Truncation of limonene synthase preprotein provides a fully active ""pseudomature"" form of this monoterpene ...
|
|
72021
|
Truncation of limonene synthase preprotein provides a fully active ""pseudomature"" form of this monoterpene ...
|
|
72022
|
Truncation of limonene synthase preprotein provides a fully active ""pseudomature"" form of this monoterpene ...
|
|
72023
|
Truncation of limonene synthase preprotein provides a fully active ""pseudomature"" form of this monoterpene ...
|
|
72024
|
Truncation of limonene synthase preprotein provides a fully active ""pseudomature"" form of this monoterpene ...
|
|
72025
|
Truncation of limonene synthase preprotein provides a fully active ""pseudomature"" form of this monoterpene ...
|
|
72026
|
Truncation of limonene synthase preprotein provides a fully active ""pseudomature"" form of this monoterpene ...
|
|
72027
|
Monoterpene synthases from common sage (Salvia officinalis). cDNA isolation, characterization, and functional ...
|
|
72028
|
Monoterpene synthases from common sage (Salvia officinalis). cDNA isolation, characterization, and functional ...
|
|
72029
|
Monoterpene synthases from common sage (Salvia officinalis). cDNA isolation, characterization, and functional ...
|
|
72030
|
Crucial role of conserved lysine 277 in the fidelity of tRNA aminoacylation by Escherichia coli valyl-tRNA ...
|
|
72031
|
Crucial role of conserved lysine 277 in the fidelity of tRNA aminoacylation by Escherichia coli valyl-tRNA ...
|
|
72032
|
Crucial role of conserved lysine 277 in the fidelity of tRNA aminoacylation by Escherichia coli valyl-tRNA ...
|
|
72033
|
Crucial role of conserved lysine 277 in the fidelity of tRNA aminoacylation by Escherichia coli valyl-tRNA ...
|
|
72034
|
Crucial role of conserved lysine 277 in the fidelity of tRNA aminoacylation by Escherichia coli valyl-tRNA ...
|
|
72035
|
Crucial role of conserved lysine 277 in the fidelity of tRNA aminoacylation by Escherichia coli valyl-tRNA ...
|
|
72036
|
Crucial role of conserved lysine 277 in the fidelity of tRNA aminoacylation by Escherichia coli valyl-tRNA ...
|
|
72037
|
Crucial role of conserved lysine 277 in the fidelity of tRNA aminoacylation by Escherichia coli valyl-tRNA ...
|
|
72038
|
Crucial role of conserved lysine 277 in the fidelity of tRNA aminoacylation by Escherichia coli valyl-tRNA ...
|
|
72039
|
Crucial role of conserved lysine 277 in the fidelity of tRNA aminoacylation by Escherichia coli valyl-tRNA ...
|
|
72040
|
Crucial role of conserved lysine 277 in the fidelity of tRNA aminoacylation by Escherichia coli valyl-tRNA ...
|
|
72041
|
Crucial role of conserved lysine 277 in the fidelity of tRNA aminoacylation by Escherichia coli valyl-tRNA ...
|
|
72042
|
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
|
|
72043
|
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
|
|
72044
|
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
|
|
72045
|
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
|
|
72046
|
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
|
|
72047
|
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
|
|
72048
|
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
|
|
72049
|
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
|
|
72050
|
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
|
|
72051
|
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
|
|
72052
|
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
|
|
72053
|
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
|
|
72054
|
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
|
|
72055
|
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
|
|
72056
|
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
|
|
72057
|
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
|
|
72058
|
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
|
|
72059
|
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
|
|
72060
|
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
|
|
72061
|
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
|
|
72062
|
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
|
|
72063
|
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
|
|
72064
|
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
|
|
72065
|
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
|
|
72066
|
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
|
|
72067
|
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
|
|
72068
|
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
|
|
72069
|
Structure and activity of the axon guidance protein MICAL.
|
|
72070
|
Structure and activity of the axon guidance protein MICAL.
|
|
72071
|
Properties and catalytic activities of MICAL1, the flavoenzyme involved in cytoskeleton dynamics, and ...
|
|
72072
|
Properties and catalytic activities of MICAL1, the flavoenzyme involved in cytoskeleton dynamics, and ...
|
|
72073
|
Properties and catalytic activities of MICAL1, the flavoenzyme involved in cytoskeleton dynamics, and ...
|
|
72074
|
Properties and catalytic activities of MICAL1, the flavoenzyme involved in cytoskeleton dynamics, and ...
|
|
72075
|
Properties and catalytic activities of MICAL1, the flavoenzyme involved in cytoskeleton dynamics, and ...
|
|
72076
|
Properties and catalytic activities of MICAL1, the flavoenzyme involved in cytoskeleton dynamics, and ...
|
|
72077
|
Properties and catalytic activities of MICAL1, the flavoenzyme involved in cytoskeleton dynamics, and ...
|
|
72078
|
Properties and catalytic activities of MICAL1, the flavoenzyme involved in cytoskeleton dynamics, and ...
|
|
72079
|
Properties and catalytic activities of MICAL1, the flavoenzyme involved in cytoskeleton dynamics, and ...
|
|
72080
|
Properties and catalytic activities of MICAL1, the flavoenzyme involved in cytoskeleton dynamics, and ...
|
|
72081
|
Properties and catalytic activities of MICAL1, the flavoenzyme involved in cytoskeleton dynamics, and ...
|
|
72082
|
Properties and catalytic activities of MICAL1, the flavoenzyme involved in cytoskeleton dynamics, and ...
|
|
72083
|
Properties and catalytic activities of MICAL1, the flavoenzyme involved in cytoskeleton dynamics, and ...
|
|
72084
|
A kinetic, spectroscopic, and redox study of human tryptophan 2,3-dioxygenase
|
|
72085
|
A kinetic, spectroscopic, and redox study of human tryptophan 2,3-dioxygenase
|
|
72086
|
A kinetic, spectroscopic, and redox study of human tryptophan 2,3-dioxygenase
|
|
72087
|
A kinetic, spectroscopic, and redox study of human tryptophan 2,3-dioxygenase
|
|
72088
|
A kinetic, spectroscopic, and redox study of human tryptophan 2,3-dioxygenase
|
|
72089
|
A kinetic, spectroscopic, and redox study of human tryptophan 2,3-dioxygenase
|
|
72090
|
A kinetic, spectroscopic, and redox study of human tryptophan 2,3-dioxygenase
|
|
72091
|
A kinetic, spectroscopic, and redox study of human tryptophan 2,3-dioxygenase
|
|
72092
|
A kinetic, spectroscopic, and redox study of human tryptophan 2,3-dioxygenase
|
|
72093
|
A kinetic, spectroscopic, and redox study of human tryptophan 2,3-dioxygenase
|
|
72094
|
A kinetic, spectroscopic, and redox study of human tryptophan 2,3-dioxygenase
|
|
72095
|
A kinetic, spectroscopic, and redox study of human tryptophan 2,3-dioxygenase
|
|
72096
|
Mutational analysis suggests the same design for editing activities of two tRNA synthetases
|
|
72097
|
Mutational analysis suggests the same design for editing activities of two tRNA synthetases
|
|
72098
|
Mutational analysis suggests the same design for editing activities of two tRNA synthetases
|
|
72099
|
Mutational analysis suggests the same design for editing activities of two tRNA synthetases
|
|
72100
|
Mutational analysis suggests the same design for editing activities of two tRNA synthetases
|
|
72101
|
Mutational analysis suggests the same design for editing activities of two tRNA synthetases
|
|
72102
|
Mutational analysis suggests the same design for editing activities of two tRNA synthetases
|
|
72103
|
Mutational analysis suggests the same design for editing activities of two tRNA synthetases
|
|
72104
|
Mutational analysis suggests the same design for editing activities of two tRNA synthetases
|
|
72105
|
Mutational analysis suggests the same design for editing activities of two tRNA synthetases
|
|
72106
|
Mutational analysis suggests the same design for editing activities of two tRNA synthetases
|
|
72107
|
Mutational analysis suggests the same design for editing activities of two tRNA synthetases
|
|
72108
|
Mutational analysis suggests the same design for editing activities of two tRNA synthetases
|
|
72109
|
Mutational analysis suggests the same design for editing activities of two tRNA synthetases
|
|
72110
|
Mutational analysis suggests the same design for editing activities of two tRNA synthetases
|
|
72111
|
A single amidotransferase forms asparaginyl-tRNA and glutaminyl-tRNA in Chlamydia trachomatis
|
|
72112
|
A single amidotransferase forms asparaginyl-tRNA and glutaminyl-tRNA in Chlamydia trachomatis
|
|
72113
|
Major anticodon-binding region missing from an archaebacterial tRNA synthetase
|
|
72114
|
Major anticodon-binding region missing from an archaebacterial tRNA synthetase
|
|
72115
|
Major anticodon-binding region missing from an archaebacterial tRNA synthetase
|
|
72116
|
Spectroscopic studies of ligand and substrate binding to human indoleamine 2,3-dioxygenase
|
|
72117
|
Spectroscopic studies of ligand and substrate binding to human indoleamine 2,3-dioxygenase
|
|
72118
|
Spectroscopic studies of ligand and substrate binding to human indoleamine 2,3-dioxygenase
|
|
72119
|
Spectroscopic studies of ligand and substrate binding to human indoleamine 2,3-dioxygenase
|
|
72120
|
Spectroscopic studies of ligand and substrate binding to human indoleamine 2,3-dioxygenase
|
|
72121
|
Spectroscopic studies of ligand and substrate binding to human indoleamine 2,3-dioxygenase
|
|
72122
|
Spectroscopic studies of ligand and substrate binding to human indoleamine 2,3-dioxygenase
|
|
72123
|
Spectroscopic studies of ligand and substrate binding to human indoleamine 2,3-dioxygenase
|
|
72124
|
Stereospecificity of hepatic L-tryptophan 2,3-dioxygenase
|
|
72125
|
Stereospecificity of hepatic L-tryptophan 2,3-dioxygenase
|
|
72126
|
Stereospecificity of hepatic L-tryptophan 2,3-dioxygenase
|
|
72127
|
Stereospecificity of hepatic L-tryptophan 2,3-dioxygenase
|
|
72128
|
Stereospecificity of hepatic L-tryptophan 2,3-dioxygenase
|
|
72129
|
Stereospecificity of hepatic L-tryptophan 2,3-dioxygenase
|
|
72130
|
Stereospecificity of hepatic L-tryptophan 2,3-dioxygenase
|
|
72131
|
Stereospecificity of hepatic L-tryptophan 2,3-dioxygenase
|
|
72132
|
Ligand and substrate migration in human indoleamine 2,3-dioxygenase
|
|
72133
|
Ligand and substrate migration in human indoleamine 2,3-dioxygenase
|
|
72134
|
Ligand and substrate migration in human indoleamine 2,3-dioxygenase
|
|
72135
|
Ligand and substrate migration in human indoleamine 2,3-dioxygenase
|
|
72136
|
Indoleamine 2,3-dioxygenase. Equilibrium studies of the tryptophan binding to the ferric, ferrous, and CO- ...
|
|
72137
|
Indoleamine 2,3-dioxygenase. Equilibrium studies of the tryptophan binding to the ferric, ferrous, and CO- ...
|
|
72138
|
Indoleamine 2,3-dioxygenase. Equilibrium studies of the tryptophan binding to the ferric, ferrous, and CO- ...
|
|
72139
|
Indoleamine 2,3-dioxygenase. Equilibrium studies of the tryptophan binding to the ferric, ferrous, and CO- ...
|
|
72140
|
Indoleamine 2,3-dioxygenase. Equilibrium studies of the tryptophan binding to the ferric, ferrous, and CO- ...
|
|
72141
|
Indoleamine 2,3-dioxygenase. Equilibrium studies of the tryptophan binding to the ferric, ferrous, and CO- ...
|
|
72142
|
Indoleamine 2,3-dioxygenase. Equilibrium studies of the tryptophan binding to the ferric, ferrous, and CO- ...
|
|
72143
|
Indoleamine 2,3-dioxygenase. Equilibrium studies of the tryptophan binding to the ferric, ferrous, and CO- ...
|
|
72144
|
Indoleamine 2,3-dioxygenase. Equilibrium studies of the tryptophan binding to the ferric, ferrous, and CO- ...
|
|
72145
|
Indoleamine 2,3-dioxygenase. Equilibrium studies of the tryptophan binding to the ferric, ferrous, and CO- ...
|
|
72146
|
Indoleamine 2,3-dioxygenase. Equilibrium studies of the tryptophan binding to the ferric, ferrous, and CO- ...
|
|
72147
|
Indoleamine 2,3-dioxygenase. Equilibrium studies of the tryptophan binding to the ferric, ferrous, and CO- ...
|
|
72148
|
Indoleamine 2,3-dioxygenase. Equilibrium studies of the tryptophan binding to the ferric, ferrous, and CO- ...
|
|
72149
|
Indoleamine 2,3-dioxygenase. Equilibrium studies of the tryptophan binding to the ferric, ferrous, and CO- ...
|
|
72150
|
Inhibition Mechanisms of Human Indoleamine 2,3 Dioxygenase 1
|
|
72151
|
Inhibition Mechanisms of Human Indoleamine 2,3 Dioxygenase 1
|
|
72152
|
Inhibition Mechanisms of Human Indoleamine 2,3 Dioxygenase 1
|
|
72153
|
Inhibition Mechanisms of Human Indoleamine 2,3 Dioxygenase 1
|
|
72154
|
Inhibition Mechanisms of Human Indoleamine 2,3 Dioxygenase 1
|
|
72155
|
Inhibition Mechanisms of Human Indoleamine 2,3 Dioxygenase 1
|
|
72156
|
Inhibition Mechanisms of Human Indoleamine 2,3 Dioxygenase 1
|
|
72157
|
Inhibition Mechanisms of Human Indoleamine 2,3 Dioxygenase 1
|
|
72158
|
Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as ...
|
|
72159
|
Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as ...
|
|
72160
|
Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as ...
|
|
72161
|
Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as ...
|
|
72162
|
Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as ...
|
|
72163
|
Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as ...
|
|
72164
|
Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as ...
|
|
72165
|
Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as ...
|
|
72166
|
Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as ...
|
|
72167
|
Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as ...
|
|
72168
|
Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as ...
|
|
72169
|
Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as ...
|
|
72170
|
Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as ...
|
|
72171
|
Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as ...
|
|
72172
|
Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as ...
|
|
72173
|
Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as ...
|
|
72174
|
Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as ...
|
|
72175
|
Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as ...
|
|
72176
|
Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as ...
|
|
72177
|
Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as ...
|
|
72178
|
Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as ...
|
|
72179
|
Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as ...
|
|
72180
|
Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as ...
|
|
72181
|
Cysteine synthase (O-acetylserine (thiol) lyase) substrate specificities classify the mitochondrial isoform as ...
|
|
72182
|
SARS-CoV 3CLpro inhibitory effects of quinone-methide triterpenes from Tripterygium regelii
|
|
72183
|
SARS-CoV 3CLpro inhibitory effects of quinone-methide triterpenes from Tripterygium regelii
|
|
72184
|
SARS-CoV 3CLpro inhibitory effects of quinone-methide triterpenes from Tripterygium regelii
|
|
72185
|
SARS-CoV 3CLpro inhibitory effects of quinone-methide triterpenes from Tripterygium regelii
|
|
72186
|
SARS-CoV 3CLpro inhibitory effects of quinone-methide triterpenes from Tripterygium regelii
|
|
72187
|
SARS-CoV 3CLpro inhibitory effects of quinone-methide triterpenes from Tripterygium regelii
|
|
72188
|
SARS-CoV 3CLpro inhibitory effects of quinone-methide triterpenes from Tripterygium regelii
|
|
72189
|
SARS-CoV 3CLpro inhibitory effects of quinone-methide triterpenes from Tripterygium regelii
|
|
72190
|
SARS-CoV 3CLpro inhibitory effects of quinone-methide triterpenes from Tripterygium regelii
|
|
72191
|
SARS-CoV 3CLpro inhibitory effects of quinone-methide triterpenes from Tripterygium regelii
|
|
72192
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72193
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72194
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72195
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72196
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72197
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72198
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72199
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72200
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72201
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72202
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72203
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72204
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72205
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72206
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72207
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72208
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72209
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72210
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72211
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72212
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72213
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72214
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72215
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72216
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72217
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72218
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72219
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72220
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72221
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72222
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72223
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72224
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72225
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72226
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72227
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72228
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72229
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72230
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72231
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72232
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72233
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72234
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72235
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72236
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72237
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72238
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72239
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72240
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72241
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72242
|
Regulation of 2-oxoglutarate metabolism in rat liver by NADP-isocitrate dehydrogenase and aspartate ...
|
|
72243
|
Evidence from cassette mutagenesis for a structure-function motif in a protein of unknown structure
|
|
72244
|
Evidence from cassette mutagenesis for a structure-function motif in a protein of unknown structure
|
|
72245
|
Evidence from cassette mutagenesis for a structure-function motif in a protein of unknown structure
|
|
72246
|
Evidence from cassette mutagenesis for a structure-function motif in a protein of unknown structure
|
|
72247
|
Evidence from cassette mutagenesis for a structure-function motif in a protein of unknown structure
|
|
72248
|
Evidence from cassette mutagenesis for a structure-function motif in a protein of unknown structure
|
|
72249
|
Virtual high- throughput screening identifies mycophenolic acid as a novel RNA capping inhibitor
|
|
72250
|
Virtual high- throughput screening identifies mycophenolic acid as a novel RNA capping inhibitor
|
|
72251
|
Virtual high- throughput screening identifies mycophenolic acid as a novel RNA capping inhibitor
|
|
72252
|
Virtual high- throughput screening identifies mycophenolic acid as a novel RNA capping inhibitor
|
|
72253
|
Free methionine-(R)-sulfoxide reductase from Escherichia coli reveals a new GAF domain function
|
|
72254
|
Free methionine-(R)-sulfoxide reductase from Escherichia coli reveals a new GAF domain function
|
|
72255
|
Parallel partial purification of cytoplasmic and mitochondrial aconitate hydratases from rat liver.
|
|
72256
|
Parallel partial purification of cytoplasmic and mitochondrial aconitate hydratases from rat liver.
|
|
72257
|
Parallel partial purification of cytoplasmic and mitochondrial aconitate hydratases from rat liver.
|
|
72258
|
Parallel partial purification of cytoplasmic and mitochondrial aconitate hydratases from rat liver.
|
|
72259
|
Purification and some properties of citrate synthase from Penicillium spiculisporum.
|
|
72260
|
Purification and some properties of citrate synthase from Penicillium spiculisporum.
|
|
72261
|
Purification and some properties of citrate synthase from Penicillium spiculisporum.
|
|
72262
|
Purification and some properties of citrate synthase from Penicillium spiculisporum.
|
|
72263
|
Purification and some properties of citrate synthase from Penicillium spiculisporum.
|
|
72264
|
Sucrose efflux mediated by SWEET proteins as a key step for phloem transport.
|
|
72265
|
Improved biomass degradation using fungal glucuronoyl-esterases-hydrolysis of natural corn fiber substrate.
|
|
72266
|
Improved biomass degradation using fungal glucuronoyl-esterases-hydrolysis of natural corn fiber substrate.
|
|
72267
|
NADP(H) phosphatase activities of archaeal inositol monophosphatase and eubacterial 3'-phosphoadenosine ...
|
|
72268
|
NADP(H) phosphatase activities of archaeal inositol monophosphatase and eubacterial 3'-phosphoadenosine ...
|
|
72269
|
NADP(H) phosphatase activities of archaeal inositol monophosphatase and eubacterial 3'-phosphoadenosine ...
|
|
72270
|
NADP(H) phosphatase activities of archaeal inositol monophosphatase and eubacterial 3'-phosphoadenosine ...
|
|
72271
|
NADP(H) phosphatase activities of archaeal inositol monophosphatase and eubacterial 3'-phosphoadenosine ...
|
|
72272
|
NADP(H) phosphatase activities of archaeal inositol monophosphatase and eubacterial 3'-phosphoadenosine ...
|
|
72273
|
NADP(H) phosphatase activities of archaeal inositol monophosphatase and eubacterial 3'-phosphoadenosine ...
|
|
72274
|
NADP(H) phosphatase activities of archaeal inositol monophosphatase and eubacterial 3'-phosphoadenosine ...
|
|
72275
|
NADP(H) phosphatase activities of archaeal inositol monophosphatase and eubacterial 3'-phosphoadenosine ...
|
|
72276
|
NADP(H) phosphatase activities of archaeal inositol monophosphatase and eubacterial 3'-phosphoadenosine ...
|
|
72277
|
NADP(H) phosphatase activities of archaeal inositol monophosphatase and eubacterial 3'-phosphoadenosine ...
|
|
72278
|
NADP(H) phosphatase activities of archaeal inositol monophosphatase and eubacterial 3'-phosphoadenosine ...
|
|
72279
|
NADP(H) phosphatase activities of archaeal inositol monophosphatase and eubacterial 3'-phosphoadenosine ...
|
|
72280
|
NADP(H) phosphatase activities of archaeal inositol monophosphatase and eubacterial 3'-phosphoadenosine ...
|
|
72281
|
NADP(H) phosphatase activities of archaeal inositol monophosphatase and eubacterial 3'-phosphoadenosine ...
|
|
72282
|
NADP(H) phosphatase activities of archaeal inositol monophosphatase and eubacterial 3'-phosphoadenosine ...
|
|
72283
|
NADP(H) phosphatase activities of archaeal inositol monophosphatase and eubacterial 3'-phosphoadenosine ...
|
|
72284
|
NADP(H) phosphatase activities of archaeal inositol monophosphatase and eubacterial 3'-phosphoadenosine ...
|
|
72285
|
NADP(H) phosphatase activities of archaeal inositol monophosphatase and eubacterial 3'-phosphoadenosine ...
|
|
72286
|
NADP(H) phosphatase activities of archaeal inositol monophosphatase and eubacterial 3'-phosphoadenosine ...
|
|
72287
|
NADP(H) phosphatase activities of archaeal inositol monophosphatase and eubacterial 3'-phosphoadenosine ...
|
|
72288
|
NADP(H) phosphatase activities of archaeal inositol monophosphatase and eubacterial 3'-phosphoadenosine ...
|
|
72289
|
NADP(H) phosphatase activities of archaeal inositol monophosphatase and eubacterial 3'-phosphoadenosine ...
|
|
72290
|
NADP(H) phosphatase activities of archaeal inositol monophosphatase and eubacterial 3'-phosphoadenosine ...
|
|
72291
|
The role of lysine residues 297 and 306 in nucleoside triphosphate regulation of E. coli CTP synthase: ...
|
|
72292
|
The role of lysine residues 297 and 306 in nucleoside triphosphate regulation of E. coli CTP synthase: ...
|
|
72293
|
The role of lysine residues 297 and 306 in nucleoside triphosphate regulation of E. coli CTP synthase: ...
|
|
72294
|
The role of lysine residues 297 and 306 in nucleoside triphosphate regulation of E. coli CTP synthase: ...
|
|
72295
|
The role of lysine residues 297 and 306 in nucleoside triphosphate regulation of E. coli CTP synthase: ...
|
|
72296
|
The role of lysine residues 297 and 306 in nucleoside triphosphate regulation of E. coli CTP synthase: ...
|
|
72297
|
The role of lysine residues 297 and 306 in nucleoside triphosphate regulation of E. coli CTP synthase: ...
|
|
72298
|
The role of lysine residues 297 and 306 in nucleoside triphosphate regulation of E. coli CTP synthase: ...
|
|
72299
|
The role of lysine residues 297 and 306 in nucleoside triphosphate regulation of E. coli CTP synthase: ...
|
|
72300
|
The role of lysine residues 297 and 306 in nucleoside triphosphate regulation of E. coli CTP synthase: ...
|
|
72301
|
The role of lysine residues 297 and 306 in nucleoside triphosphate regulation of E. coli CTP synthase: ...
|
|
72302
|
The role of lysine residues 297 and 306 in nucleoside triphosphate regulation of E. coli CTP synthase: ...
|
|
72303
|
The role of lysine residues 297 and 306 in nucleoside triphosphate regulation of E. coli CTP synthase: ...
|
|
72304
|
The role of lysine residues 297 and 306 in nucleoside triphosphate regulation of E. coli CTP synthase: ...
|
|
72305
|
The role of lysine residues 297 and 306 in nucleoside triphosphate regulation of E. coli CTP synthase: ...
|
|
72306
|
The role of lysine residues 297 and 306 in nucleoside triphosphate regulation of E. coli CTP synthase: ...
|
|
72307
|
The role of lysine residues 297 and 306 in nucleoside triphosphate regulation of E. coli CTP synthase: ...
|
|
72308
|
The role of lysine residues 297 and 306 in nucleoside triphosphate regulation of E. coli CTP synthase: ...
|
|
72309
|
The role of lysine residues 297 and 306 in nucleoside triphosphate regulation of E. coli CTP synthase: ...
|
|
72310
|
The role of lysine residues 297 and 306 in nucleoside triphosphate regulation of E. coli CTP synthase: ...
|
|
72311
|
The role of lysine residues 297 and 306 in nucleoside triphosphate regulation of E. coli CTP synthase: ...
|
|
72312
|
The role of lysine residues 297 and 306 in nucleoside triphosphate regulation of E. coli CTP synthase: ...
|
|
72313
|
The structure and mechanism of the Mycobacterium tuberculosis cyclodityrosine synthetase
|
|
72314
|
A high-throughput FRET-based assay for determination of Atg4 activity
|
|
72315
|
A high-throughput FRET-based assay for determination of Atg4 activity
|
|
72316
|
A high-throughput FRET-based assay for determination of Atg4 activity
|
|
72317
|
A high-throughput FRET-based assay for determination of Atg4 activity
|
|
72318
|
A high-throughput FRET-based assay for determination of Atg4 activity
|
|
72319
|
A high-throughput FRET-based assay for determination of Atg4 activity
|
|
72320
|
WrbA from Escherichia coli and Archaeoglobus fulgidus is an NAD(P)H:quinone oxidoreductase.
|
|
72321
|
WrbA from Escherichia coli and Archaeoglobus fulgidus is an NAD(P)H:quinone oxidoreductase.
|
|
72322
|
WrbA from Escherichia coli and Archaeoglobus fulgidus is an NAD(P)H:quinone oxidoreductase.
|
|
72323
|
WrbA from Escherichia coli and Archaeoglobus fulgidus is an NAD(P)H:quinone oxidoreductase.
|
|
72324
|
WrbA from Escherichia coli and Archaeoglobus fulgidus is an NAD(P)H:quinone oxidoreductase.
|
|
72325
|
WrbA from Escherichia coli and Archaeoglobus fulgidus is an NAD(P)H:quinone oxidoreductase.
|
|
72326
|
Identification of an osmo-dependent and an osmo-independent choline transporter in Acinetobacter baylyi: ...
|
|
72327
|
Identification of an osmo-dependent and an osmo-independent choline transporter in Acinetobacter baylyi: ...
|
|
72328
|
Impact of the lanthanide contraction on the activity of a lanthanide-dependent methanol dehydrogenase - a ...
|
|
72329
|
Impact of the lanthanide contraction on the activity of a lanthanide-dependent methanol dehydrogenase - a ...
|
|
72330
|
Impact of the lanthanide contraction on the activity of a lanthanide-dependent methanol dehydrogenase - a ...
|
|
72331
|
Impact of the lanthanide contraction on the activity of a lanthanide-dependent methanol dehydrogenase - a ...
|
|
72332
|
Impact of the lanthanide contraction on the activity of a lanthanide-dependent methanol dehydrogenase - a ...
|
|
72333
|
Impact of the lanthanide contraction on the activity of a lanthanide-dependent methanol dehydrogenase - a ...
|
|
72334
|
Impact of the lanthanide contraction on the activity of a lanthanide-dependent methanol dehydrogenase - a ...
|
|
72335
|
A genetic mechanism for Tibetan high-altitude adaptation.
|
|
72336
|
A genetic mechanism for Tibetan high-altitude adaptation.
|
|
72337
|
A genetic mechanism for Tibetan high-altitude adaptation.
|
|
72338
|
A genetic mechanism for Tibetan high-altitude adaptation.
|
|
72339
|
A genetic mechanism for Tibetan high-altitude adaptation.
|
|
72340
|
A genetic mechanism for Tibetan high-altitude adaptation.
|
|
72341
|
A genetic mechanism for Tibetan high-altitude adaptation.
|
|
72342
|
A genetic mechanism for Tibetan high-altitude adaptation.
|
|
72343
|
Ptc7p Dephosphorylates Select Mitochondrial Proteins to Enhance Metabolic Function.
|
|
72344
|
Ptc7p Dephosphorylates Select Mitochondrial Proteins to Enhance Metabolic Function.
|
|
72345
|
Ptc7p Dephosphorylates Select Mitochondrial Proteins to Enhance Metabolic Function.
|
|
72346
|
Ptc7p Dephosphorylates Select Mitochondrial Proteins to Enhance Metabolic Function.
|
|
72347
|
Ptc7p Dephosphorylates Select Mitochondrial Proteins to Enhance Metabolic Function.
|
|
72348
|
Ptc7p Dephosphorylates Select Mitochondrial Proteins to Enhance Metabolic Function.
|
|
72349
|
Development of a red-shifted fluorescence-based assay for SARS-coronavirus 3CL protease: identification of a ...
|
|
72350
|
Development of a red-shifted fluorescence-based assay for SARS-coronavirus 3CL protease: identification of a ...
|
|
72351
|
Development of a red-shifted fluorescence-based assay for SARS-coronavirus 3CL protease: identification of a ...
|
|
72352
|
Development of a red-shifted fluorescence-based assay for SARS-coronavirus 3CL protease: identification of a ...
|
|
72353
|
Two active forms of UDP-N-acetylglucosamine enolpyruvyl transferase in gram-positive bacteria
|
|
72354
|
Two active forms of UDP-N-acetylglucosamine enolpyruvyl transferase in gram-positive bacteria
|
|
72355
|
Two active forms of UDP-N-acetylglucosamine enolpyruvyl transferase in gram-positive bacteria
|
|
72356
|
Two active forms of UDP-N-acetylglucosamine enolpyruvyl transferase in gram-positive bacteria
|
|
72357
|
Discovery of N-(1-(3-fluorobenzoyl)-1H-indol-5-yl)pyrazine-2-carboxamide: a novel, selective, ...
|
|
72358
|
Discovery of N-(1-(3-fluorobenzoyl)-1H-indol-5-yl)pyrazine-2-carboxamide: a novel, selective, ...
|
|
72359
|
Discovery of N-(1-(3-fluorobenzoyl)-1H-indol-5-yl)pyrazine-2-carboxamide: a novel, selective, ...
|
|
72360
|
Purification of rat heart and rat liver citrate synthases. Physical, kinetic, and immunological studies.
|
|
72361
|
Purification of rat heart and rat liver citrate synthases. Physical, kinetic, and immunological studies.
|
|
72362
|
Purification of rat heart and rat liver citrate synthases. Physical, kinetic, and immunological studies.
|
|
72363
|
Purification of rat heart and rat liver citrate synthases. Physical, kinetic, and immunological studies.
|
|
72364
|
Purification of rat heart and rat liver citrate synthases. Physical, kinetic, and immunological studies.
|
|
72365
|
Purification of rat heart and rat liver citrate synthases. Physical, kinetic, and immunological studies.
|
|
72366
|
The fungal product terreic acid is a covalent inhibitor of the bacterial cell wall biosynthetic enzyme ...
|
|
72367
|
The fungal product terreic acid is a covalent inhibitor of the bacterial cell wall biosynthetic enzyme ...
|
|
72368
|
The fungal product terreic acid is a covalent inhibitor of the bacterial cell wall biosynthetic enzyme ...
|
|
72369
|
The fungal product terreic acid is a covalent inhibitor of the bacterial cell wall biosynthetic enzyme ...
|
|
72370
|
The fungal product terreic acid is a covalent inhibitor of the bacterial cell wall biosynthetic enzyme ...
|
|
72371
|
The fungal product terreic acid is a covalent inhibitor of the bacterial cell wall biosynthetic enzyme ...
|
|
72372
|
CYP15A1, the cytochrome P450 that catalyzes epoxidation of methyl farnesoate to juvenile hormone III in ...
|
|
72373
|
CYP15A1, the cytochrome P450 that catalyzes epoxidation of methyl farnesoate to juvenile hormone III in ...
|
|
72374
|
CYP15A1, the cytochrome P450 that catalyzes epoxidation of methyl farnesoate to juvenile hormone III in ...
|
|
72375
|
CYP15A1, the cytochrome P450 that catalyzes epoxidation of methyl farnesoate to juvenile hormone III in ...
|
|
72376
|
CYP15A1, the cytochrome P450 that catalyzes epoxidation of methyl farnesoate to juvenile hormone III in ...
|
|
72377
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72378
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72379
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72380
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72381
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72382
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72383
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72384
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72385
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72386
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72387
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72388
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72389
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72390
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72391
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72392
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72393
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72394
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72395
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72396
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72397
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72398
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72399
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72400
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72401
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72402
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72403
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72404
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72405
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72406
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72407
|
The stratagem utilized by the plasminogen activator from the snake Trimeresurus stejnegeri to escape serpins
|
|
72408
|
Characterization of a new platelet aggregating factor from crotoxin Crotalus durissus cascavella venom
|
|
72409
|
Actions of two serine proteases from Trimeresurus jerdonii venom on chromogenic substrates and fibrinogen
|
|
72410
|
Actions of two serine proteases from Trimeresurus jerdonii venom on chromogenic substrates and fibrinogen
|
|
72411
|
Actions of two serine proteases from Trimeresurus jerdonii venom on chromogenic substrates and fibrinogen
|
|
72412
|
Actions of two serine proteases from Trimeresurus jerdonii venom on chromogenic substrates and fibrinogen
|
|
72413
|
Actions of two serine proteases from Trimeresurus jerdonii venom on chromogenic substrates and fibrinogen
|
|
72414
|
Characterization of a fibrinogen-clotting enzyme from Trimeresurus stejnegeri venom, and comparative study ...
|
|
72415
|
Characterization of a fibrinogen-clotting enzyme from Trimeresurus stejnegeri venom, and comparative study ...
|
|
72416
|
Characterization of a fibrinogen-clotting enzyme from Trimeresurus stejnegeri venom, and comparative study ...
|
|
72417
|
Characterization of a fibrinogen-clotting enzyme from Trimeresurus stejnegeri venom, and comparative study ...
|
|
72418
|
Characterization of a fibrinogen-clotting enzyme from Trimeresurus stejnegeri venom, and comparative study ...
|
|
72419
|
Characterization of a fibrinogen-clotting enzyme from Trimeresurus stejnegeri venom, and comparative study ...
|
|
72420
|
Characterization of a fibrinogen-clotting enzyme from Trimeresurus stejnegeri venom, and comparative study ...
|
|
72421
|
Characterization of a fibrinogen-clotting enzyme from Trimeresurus stejnegeri venom, and comparative study ...
|
|
72422
|
Characterization of a fibrinogen-clotting enzyme from Trimeresurus stejnegeri venom, and comparative study ...
|
|
72423
|
Characterization of a fibrinogen-clotting enzyme from Trimeresurus stejnegeri venom, and comparative study ...
|
|
72424
|
Characterization of a fibrinogen-clotting enzyme from Trimeresurus stejnegeri venom, and comparative study ...
|
|
72425
|
Characterization of a fibrinogen-clotting enzyme from Trimeresurus stejnegeri venom, and comparative study ...
|
|
72426
|
Characterization of a fibrinogen-clotting enzyme from Trimeresurus stejnegeri venom, and comparative study ...
|
|
72427
|
Characterization of a fibrinogen-clotting enzyme from Trimeresurus stejnegeri venom, and comparative study ...
|
|
72428
|
Characterization of a fibrinogen-clotting enzyme from Trimeresurus stejnegeri venom, and comparative study ...
|
|
72429
|
Characterization of a fibrinogen-clotting enzyme from Trimeresurus stejnegeri venom, and comparative study ...
|
|
72430
|
Characterization of a fibrinogen-clotting enzyme from Trimeresurus stejnegeri venom, and comparative study ...
|
|
72431
|
Characterization of a fibrinogen-clotting enzyme from Trimeresurus stejnegeri venom, and comparative study ...
|
|
72432
|
Characterization of a fibrinogen-clotting enzyme from Trimeresurus stejnegeri venom, and comparative study ...
|
|
72433
|
Characterization of a fibrinogen-clotting enzyme from Trimeresurus stejnegeri venom, and comparative study ...
|
|
72434
|
Characterization of a fibrinogen-clotting enzyme from Trimeresurus stejnegeri venom, and comparative study ...
|
|
72435
|
Characterization of three fibrinogenolytic enzymes from Chinese green tree viper (Trimeresurus stejnegeri) ...
|
|
72436
|
Characterization of three fibrinogenolytic enzymes from Chinese green tree viper (Trimeresurus stejnegeri) ...
|
|
72437
|
Characterization of three fibrinogenolytic enzymes from Chinese green tree viper (Trimeresurus stejnegeri) ...
|
|
72438
|
Characterization of three fibrinogenolytic enzymes from Chinese green tree viper (Trimeresurus stejnegeri) ...
|
|
72439
|
Characterization of three fibrinogenolytic enzymes from Chinese green tree viper (Trimeresurus stejnegeri) ...
|
|
72440
|
Characterization of three fibrinogenolytic enzymes from Chinese green tree viper (Trimeresurus stejnegeri) ...
|
|
72441
|
Characterization of three fibrinogenolytic enzymes from Chinese green tree viper (Trimeresurus stejnegeri) ...
|
|
72442
|
Characterization of three fibrinogenolytic enzymes from Chinese green tree viper (Trimeresurus stejnegeri) ...
|
|
72443
|
Characterization of three fibrinogenolytic enzymes from Chinese green tree viper (Trimeresurus stejnegeri) ...
|
|
72444
|
Characterization of three fibrinogenolytic enzymes from Chinese green tree viper (Trimeresurus stejnegeri) ...
|
|
72445
|
Characterization of three fibrinogenolytic enzymes from Chinese green tree viper (Trimeresurus stejnegeri) ...
|
|
72446
|
Characterization of three fibrinogenolytic enzymes from Chinese green tree viper (Trimeresurus stejnegeri) ...
|
|
72447
|
Characterization of three fibrinogenolytic enzymes from Chinese green tree viper (Trimeresurus stejnegeri) ...
|
|
72448
|
Characterization of three fibrinogenolytic enzymes from Chinese green tree viper (Trimeresurus stejnegeri) ...
|
|
72449
|
Characterization of three fibrinogenolytic enzymes from Chinese green tree viper (Trimeresurus stejnegeri) ...
|
|
72450
|
Biochemical and biological characterization of two serine proteinases from Colombian Crotalus durissus ...
|
|
72451
|
Biochemical and biological characterization of two serine proteinases from Colombian Crotalus durissus ...
|
|
72452
|
Biochemical characterization and molecular cloning of a plasminogen activator proteinase (LV-PA) from ...
|
|
72453
|
Biochemical characterization and molecular cloning of a plasminogen activator proteinase (LV-PA) from ...
|
|
72454
|
Biochemical characterization and molecular cloning of a plasminogen activator proteinase (LV-PA) from ...
|
|
72455
|
Biochemical characterization and molecular cloning of a plasminogen activator proteinase (LV-PA) from ...
|
|
72456
|
Beta-fibrinogenase from the venom of Vipera lebetina
|
|
72457
|
Beta-fibrinogenase from the venom of Vipera lebetina
|
|
72458
|
Beta-fibrinogenase from the venom of Vipera lebetina
|
|
72459
|
Jerdonase, a novel serine protease with kinin-releasing and fibrinogenolytic activity from Trimeresurus ...
|
|
72460
|
Jerdonase, a novel serine protease with kinin-releasing and fibrinogenolytic activity from Trimeresurus ...
|
|
72461
|
Isolation and characterization of the thrombin-like enzyme from Cryptelytrops albolabris (white-lipped tree ...
|
|
72462
|
Isolation and characterization of the thrombin-like enzyme from Cryptelytrops albolabris (white-lipped tree ...
|
|
72463
|
Isolation and characterization of the thrombin-like enzyme from Cryptelytrops albolabris (white-lipped tree ...
|
|
72464
|
Isolation and characterization of the thrombin-like enzyme from Cryptelytrops albolabris (white-lipped tree ...
|
|
72465
|
Isolation and characterization of the thrombin-like enzyme from Cryptelytrops albolabris (white-lipped tree ...
|
|
72466
|
Kallidin-releasing enzyme from Bitis arietans (puff adder) venom
|
|
72467
|
Kallidin-releasing enzyme from Bitis arietans (puff adder) venom
|
|
72468
|
Isolation and characterization of a protein C activator from tropical moccasin venom
|
|
72469
|
Isolation and characterization of a protein C activator from tropical moccasin venom
|
|
72470
|
Isolation and characterization of a protein C activator from tropical moccasin venom
|
|
72471
|
Isolation and characterization of a protein C activator from tropical moccasin venom
|
|
72472
|
Isolation and characterization of a protein C activator from tropical moccasin venom
|
|
72473
|
Isolation and characterization of a protein C activator from tropical moccasin venom
|
|
72474
|
Isolation and characterization of a protein C activator from tropical moccasin venom
|
|
72475
|
Isolation and characterization of a protein C activator from tropical moccasin venom
|
|
72476
|
Isolation and characterization of a protein C activator from tropical moccasin venom
|
|
72477
|
Isolation and characterization of a protein C activator from tropical moccasin venom
|
|
72478
|
Isolation and characterization of a protein C activator from tropical moccasin venom
|
|
72479
|
Isolation and characterization of a protein C activator from tropical moccasin venom
|
|
72480
|
Isolation and characterization of a protein C activator from tropical moccasin venom
|
|
72481
|
A prothrombin activator serine protease from the Lonomia obliqua caterpillar venom (Lopap) biochemical ...
|
|
72482
|
Functional analysis of the transmembrane domain and activation cleavage of human corin: design and ...
|
|
72483
|
Functional analysis of the transmembrane domain and activation cleavage of human corin: design and ...
|
|
72484
|
Functional analysis of the transmembrane domain and activation cleavage of human corin: design and ...
|
|
72485
|
Functional analysis of the transmembrane domain and activation cleavage of human corin: design and ...
|
|
72486
|
Functional analysis of the transmembrane domain and activation cleavage of human corin: design and ...
|
|
72487
|
Inhibitory mechanism of novel inhibitors of UDP-N-acetylglucosamine enolpyruvyl transferase from Haemophilus ...
|
|
72488
|
Inhibitory mechanism of novel inhibitors of UDP-N-acetylglucosamine enolpyruvyl transferase from Haemophilus ...
|
|
72489
|
Inhibitory mechanism of novel inhibitors of UDP-N-acetylglucosamine enolpyruvyl transferase from Haemophilus ...
|
|
72490
|
Inhibitory mechanism of novel inhibitors of UDP-N-acetylglucosamine enolpyruvyl transferase from Haemophilus ...
|
|
72491
|
Purification and characterization of a coagulant enzyme, okinaxobin II, from Trimeresurus okinavensis ...
|
|
72492
|
Purification and characterization of a coagulant enzyme, okinaxobin II, from Trimeresurus okinavensis ...
|
|
72493
|
Purification and characterization of a coagulant enzyme, okinaxobin II, from Trimeresurus okinavensis ...
|
|
72494
|
Purification and characterization of a coagulant enzyme, okinaxobin II, from Trimeresurus okinavensis ...
|
|
72495
|
Purification and characterization of a coagulant enzyme, okinaxobin II, from Trimeresurus okinavensis ...
|
|
72496
|
Purification and characterization of a coagulant enzyme, okinaxobin II, from Trimeresurus okinavensis ...
|
|
72497
|
Biochemical and biophysical studies of Helicobacter pylori arginine decarboxylase, an enzyme important for ...
|
|
72498
|
Biochemical and biophysical studies of Helicobacter pylori arginine decarboxylase, an enzyme important for ...
|
|
72499
|
Molecular cloning and functional characterization of psoralen synthase, the first committed monooxygenase of ...
|
|
72500
|
Molecular cloning and functional characterization of psoralen synthase, the first committed monooxygenase of ...
|
|
72501
|
Molecular cloning and functional characterization of psoralen synthase, the first committed monooxygenase of ...
|
|
72502
|
Molecular cloning and functional characterization of psoralen synthase, the first committed monooxygenase of ...
|
|
72503
|
Characterization of the kaurene oxidase CYP701A3, a multifunctional cytochrome P450 from gibberellin ...
|
|
72504
|
Characterization of the kaurene oxidase CYP701A3, a multifunctional cytochrome P450 from gibberellin ...
|
|
72505
|
Characterization of the kaurene oxidase CYP701A3, a multifunctional cytochrome P450 from gibberellin ...
|
|
72506
|
Characterization of the kaurene oxidase CYP701A3, a multifunctional cytochrome P450 from gibberellin ...
|
|
72507
|
Characterization of the kaurene oxidase CYP701A3, a multifunctional cytochrome P450 from gibberellin ...
|
|
72508
|
Sulfonamido, azidosulfonyl and N-acetylsulfonamido analogues of rofecoxib: ...
|
|
72509
|
Sulfonamido, azidosulfonyl and N-acetylsulfonamido analogues of rofecoxib: ...
|
|
72510
|
Sulfonamido, azidosulfonyl and N-acetylsulfonamido analogues of rofecoxib: ...
|
|
72511
|
Sulfonamido, azidosulfonyl and N-acetylsulfonamido analogues of rofecoxib: ...
|
|
72512
|
Sulfonamido, azidosulfonyl and N-acetylsulfonamido analogues of rofecoxib: ...
|
|
72513
|
Sulfonamido, azidosulfonyl and N-acetylsulfonamido analogues of rofecoxib: ...
|
|
72514
|
Sulfonamido, azidosulfonyl and N-acetylsulfonamido analogues of rofecoxib: ...
|
|
72515
|
Study of tyrosine and dopa enantiomers as tyrosinase substrates initiating l- and d-melanogenesis pathways.
|
|
72516
|
Study of tyrosine and dopa enantiomers as tyrosinase substrates initiating l- and d-melanogenesis pathways.
|
|
72517
|
Study of tyrosine and dopa enantiomers as tyrosinase substrates initiating l- and d-melanogenesis pathways.
|
|
72518
|
Study of tyrosine and dopa enantiomers as tyrosinase substrates initiating l- and d-melanogenesis pathways.
|
|
72519
|
Kinetic, spectral, and structural studies of the slow-binding inhibition of the Escherichia coli ...
|
|
72520
|
Kinetic, spectral, and structural studies of the slow-binding inhibition of the Escherichia coli ...
|
|
72521
|
Mechanistic analysis of the argE-encoded N-acetylornithine deacetylase
|
|
72522
|
Mechanistic analysis of the argE-encoded N-acetylornithine deacetylase
|
|
72523
|
Mechanistic analysis of the argE-encoded N-acetylornithine deacetylase
|
|
72524
|
Mechanistic analysis of the argE-encoded N-acetylornithine deacetylase
|
|
72525
|
Mechanistic analysis of the argE-encoded N-acetylornithine deacetylase
|
|
72526
|
Mechanistic analysis of the argE-encoded N-acetylornithine deacetylase
|
|
72527
|
Mechanistic analysis of the argE-encoded N-acetylornithine deacetylase
|
|
72528
|
Mechanistic analysis of the argE-encoded N-acetylornithine deacetylase
|
|
72529
|
Mechanistic analysis of the argE-encoded N-acetylornithine deacetylase
|
|
72530
|
Mechanistic analysis of the argE-encoded N-acetylornithine deacetylase
|
|
72531
|
Mechanistic analysis of the argE-encoded N-acetylornithine deacetylase
|
|
72532
|
Mechanistic analysis of the argE-encoded N-acetylornithine deacetylase
|
|
72533
|
Mechanistic analysis of the argE-encoded N-acetylornithine deacetylase
|
|
72534
|
Mechanistic analysis of the argE-encoded N-acetylornithine deacetylase
|
|
72535
|
Mechanistic analysis of the argE-encoded N-acetylornithine deacetylase
|
|
72536
|
Mechanistic analysis of the argE-encoded N-acetylornithine deacetylase
|
|
72537
|
Mechanistic analysis of the argE-encoded N-acetylornithine deacetylase
|
|
72538
|
Mechanistic analysis of the argE-encoded N-acetylornithine deacetylase
|
|
72539
|
Mechanistic analysis of the argE-encoded N-acetylornithine deacetylase
|
|
72540
|
Interconversion of carbamayl-L-aspartate and L-dihydroorotate by dihydroorotase from mouse Ehrlich ascites ...
|
|
72541
|
3-Keto-5alpha-steroid Delta(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in ...
|
|
72542
|
3-Keto-5alpha-steroid Delta(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in ...
|
|
72543
|
3-Keto-5alpha-steroid Delta(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in ...
|
|
72544
|
3-Keto-5alpha-steroid Delta(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in ...
|
|
72545
|
3-Keto-5alpha-steroid Delta(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in ...
|
|
72546
|
3-Keto-5alpha-steroid Delta(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in ...
|
|
72547
|
3-Keto-5alpha-steroid Delta(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in ...
|
|
72548
|
3-Keto-5alpha-steroid Delta(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in ...
|
|
72549
|
3-Keto-5alpha-steroid Delta(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in ...
|
|
72550
|
3-Keto-5alpha-steroid Delta(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in ...
|
|
72551
|
3-Keto-5alpha-steroid Delta(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in ...
|
|
72552
|
3-Keto-5alpha-steroid Delta(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in ...
|
|
72553
|
3-Keto-5alpha-steroid Delta(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in ...
|
|
72554
|
3-Keto-5alpha-steroid Delta(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in ...
|
|
72555
|
3-Keto-5alpha-steroid Delta(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in ...
|
|
72556
|
3-Keto-5alpha-steroid Delta(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in ...
|
|
72557
|
3-Keto-5alpha-steroid Delta(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in ...
|
|
72558
|
3-Keto-5alpha-steroid Delta(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in ...
|
|
72559
|
3-Keto-5alpha-steroid Delta(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in ...
|
|
72560
|
3-Keto-5alpha-steroid Delta(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in ...
|
|
72561
|
3-Keto-5alpha-steroid Delta(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in ...
|
|
72562
|
3-Keto-5alpha-steroid Delta(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in ...
|
|
72563
|
3-Keto-5alpha-steroid Delta(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in ...
|
|
72564
|
Characterization of wild-type and mutant forms of human tryptophan hydroxylase 2
|
|
72565
|
Characterization of wild-type and mutant forms of human tryptophan hydroxylase 2
|
|
72566
|
Characterization of wild-type and mutant forms of human tryptophan hydroxylase 2
|
|
72567
|
Characterization of wild-type and mutant forms of human tryptophan hydroxylase 2
|
|
72568
|
Characterization of wild-type and mutant forms of human tryptophan hydroxylase 2
|
|
72569
|
Characterization of wild-type and mutant forms of human tryptophan hydroxylase 2
|
|
72570
|
Characterization of wild-type and mutant forms of human tryptophan hydroxylase 2
|
|
72571
|
Characterization of wild-type and mutant forms of human tryptophan hydroxylase 2
|
|
72572
|
Characterization of wild-type and mutant forms of human tryptophan hydroxylase 2
|
|
72573
|
Characterization of wild-type and mutant forms of human tryptophan hydroxylase 2
|
|
72574
|
Characterization of wild-type and mutant forms of human tryptophan hydroxylase 2
|
|
72575
|
Characterization of wild-type and mutant forms of human tryptophan hydroxylase 2
|
|
72576
|
Characterization of wild-type and mutant forms of human tryptophan hydroxylase 2
|
|
72577
|
Characterization of wild-type and mutant forms of human tryptophan hydroxylase 2
|
|
72578
|
Characterization of wild-type and mutant forms of human tryptophan hydroxylase 2
|
|
72579
|
Characterization of wild-type and mutant forms of human tryptophan hydroxylase 2
|
|
72580
|
Characterization of wild-type and mutant forms of human tryptophan hydroxylase 2
|
|
72581
|
Expression, purification and enzymatic characterization of the catalytic domains of human tryptophan ...
|
|
72582
|
Expression, purification and enzymatic characterization of the catalytic domains of human tryptophan ...
|
|
72583
|
Expression, purification and enzymatic characterization of the catalytic domains of human tryptophan ...
|
|
72584
|
Expression, purification and enzymatic characterization of the catalytic domains of human tryptophan ...
|
|
72585
|
Expression, purification and enzymatic characterization of the catalytic domains of human tryptophan ...
|
|
72586
|
Expression, purification and enzymatic characterization of the catalytic domains of human tryptophan ...
|
|
72587
|
Functional domains of human tryptophan hydroxylase 2 (hTPH2)
|
|
72588
|
Functional domains of human tryptophan hydroxylase 2 (hTPH2)
|
|
72589
|
Functional domains of human tryptophan hydroxylase 2 (hTPH2)
|
|
72590
|
Functional domains of human tryptophan hydroxylase 2 (hTPH2)
|
|
72591
|
Functional domains of human tryptophan hydroxylase 2 (hTPH2)
|
|
72592
|
Functional domains of human tryptophan hydroxylase 2 (hTPH2)
|
|
72593
|
Activation and stabilization of human tryptophan hydroxylase 2 by phosphorylation and 14-3-3 binding.
|
|
72594
|
Activation and stabilization of human tryptophan hydroxylase 2 by phosphorylation and 14-3-3 binding.
|
|
72595
|
Activation and stabilization of human tryptophan hydroxylase 2 by phosphorylation and 14-3-3 binding.
|
|
72596
|
Activation and stabilization of human tryptophan hydroxylase 2 by phosphorylation and 14-3-3 binding.
|
|
72597
|
Activation and stabilization of human tryptophan hydroxylase 2 by phosphorylation and 14-3-3 binding.
|
|
72598
|
Activation and stabilization of human tryptophan hydroxylase 2 by phosphorylation and 14-3-3 binding.
|
|
72599
|
Activation and stabilization of human tryptophan hydroxylase 2 by phosphorylation and 14-3-3 binding.
|
|
72600
|
Activation and stabilization of human tryptophan hydroxylase 2 by phosphorylation and 14-3-3 binding.
|
|
72601
|
Activation and stabilization of human tryptophan hydroxylase 2 by phosphorylation and 14-3-3 binding.
|
|
72602
|
Activation and stabilization of human tryptophan hydroxylase 2 by phosphorylation and 14-3-3 binding.
|
|
72603
|
Activation and stabilization of human tryptophan hydroxylase 2 by phosphorylation and 14-3-3 binding.
|
|
72604
|
Activation and stabilization of human tryptophan hydroxylase 2 by phosphorylation and 14-3-3 binding.
|
|
72605
|
Activation and stabilization of human tryptophan hydroxylase 2 by phosphorylation and 14-3-3 binding.
|
|
72606
|
Activation and stabilization of human tryptophan hydroxylase 2 by phosphorylation and 14-3-3 binding.
|
|
72607
|
Activation and stabilization of human tryptophan hydroxylase 2 by phosphorylation and 14-3-3 binding.
|
|
72608
|
Activation and stabilization of human tryptophan hydroxylase 2 by phosphorylation and 14-3-3 binding.
|
|
72609
|
Activation and stabilization of human tryptophan hydroxylase 2 by phosphorylation and 14-3-3 binding.
|
|
72610
|
Activation and stabilization of human tryptophan hydroxylase 2 by phosphorylation and 14-3-3 binding.
|
|
72611
|
Activation and stabilization of human tryptophan hydroxylase 2 by phosphorylation and 14-3-3 binding.
|
|
72612
|
Activation and stabilization of human tryptophan hydroxylase 2 by phosphorylation and 14-3-3 binding.
|
|
72613
|
Inhibition by the branched-chain 2-oxo acids of the 2-oxoglutarate dehydrogenase complex in developing rat and ...
|
|
72614
|
Inhibition by the branched-chain 2-oxo acids of the 2-oxoglutarate dehydrogenase complex in developing rat and ...
|
|
72615
|
Inhibition by the branched-chain 2-oxo acids of the 2-oxoglutarate dehydrogenase complex in developing rat and ...
|
|
72616
|
Inhibition by the branched-chain 2-oxo acids of the 2-oxoglutarate dehydrogenase complex in developing rat and ...
|
|
72617
|
Inhibition by the branched-chain 2-oxo acids of the 2-oxoglutarate dehydrogenase complex in developing rat and ...
|
|
72618
|
Inhibition by the branched-chain 2-oxo acids of the 2-oxoglutarate dehydrogenase complex in developing rat and ...
|
|
72619
|
Inhibition by the branched-chain 2-oxo acids of the 2-oxoglutarate dehydrogenase complex in developing rat and ...
|
|
72620
|
Inhibition by the branched-chain 2-oxo acids of the 2-oxoglutarate dehydrogenase complex in developing rat and ...
|
|
72621
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72622
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72623
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72624
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72625
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72626
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72627
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72628
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72629
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72630
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72631
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72632
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72633
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72634
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72635
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72636
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72637
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72638
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72639
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72640
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72641
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72642
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72643
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72644
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72645
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72646
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72647
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72648
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72649
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72650
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72651
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72652
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72653
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72654
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72655
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72656
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72657
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72658
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72659
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72660
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72661
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72662
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72663
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72664
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72665
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72666
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72667
|
Relationship of human liver dihydrodiol dehydrogenases to hepatic bile-acid-binding protein and an ...
|
|
72668
|
Substrate Oxidation by Indoleamine 2,3-Dioxygenase: EVIDENCE FOR A COMMON REACTION MECHANISM
|
|
72669
|
Catalytic synergy in the multifunctional protein that initiates pyrimidine biosynthesis in Syrian hamster ...
|
|
72670
|
Catalytic synergy in the multifunctional protein that initiates pyrimidine biosynthesis in Syrian hamster ...
|
|
72671
|
Catalytic synergy in the multifunctional protein that initiates pyrimidine biosynthesis in Syrian hamster ...
|
|
72672
|
Catalytic synergy in the multifunctional protein that initiates pyrimidine biosynthesis in Syrian hamster ...
|
|
72673
|
Catalytic synergy in the multifunctional protein that initiates pyrimidine biosynthesis in Syrian hamster ...
|
|
72674
|
Catalytic synergy in the multifunctional protein that initiates pyrimidine biosynthesis in Syrian hamster ...
|
|
72675
|
Catalytic synergy in the multifunctional protein that initiates pyrimidine biosynthesis in Syrian hamster ...
|
|
72676
|
The dihydroorotase domain of the multifunctional protein CAD. Subunit structure, zinc content, and kinetics
|
|
72677
|
The dihydroorotase domain of the multifunctional protein CAD. Subunit structure, zinc content, and kinetics
|
|
72678
|
Substitutions in the aspartate transcarbamoylase domain of hamster CAD disrupt oligomeric structure
|
|
72679
|
Substitutions in the aspartate transcarbamoylase domain of hamster CAD disrupt oligomeric structure
|
|
72680
|
Substitutions in the aspartate transcarbamoylase domain of hamster CAD disrupt oligomeric structure
|
|
72681
|
The effects of pH and inhibitors upon the catalytic activity of the dihydroorotase of multienzymatic protein ...
|
|
72682
|
The effects of pH and inhibitors upon the catalytic activity of the dihydroorotase of multienzymatic protein ...
|
|
72683
|
The effects of pH and inhibitors upon the catalytic activity of the dihydroorotase of multienzymatic protein ...
|
|
72684
|
The effects of pH and inhibitors upon the catalytic activity of the dihydroorotase of multienzymatic protein ...
|
|
72685
|
The effects of pH and inhibitors upon the catalytic activity of the dihydroorotase of multienzymatic protein ...
|
|
72686
|
The effects of pH and inhibitors upon the catalytic activity of the dihydroorotase of multienzymatic protein ...
|
|
72687
|
The effects of pH and inhibitors upon the catalytic activity of the dihydroorotase of multienzymatic protein ...
|
|
72688
|
The effects of pH and inhibitors upon the catalytic activity of the dihydroorotase of multienzymatic protein ...
|
|
72689
|
The effects of pH and inhibitors upon the catalytic activity of the dihydroorotase of multienzymatic protein ...
|
|
72690
|
The effects of pH and inhibitors upon the catalytic activity of the dihydroorotase of multienzymatic protein ...
|
|
72691
|
The effects of pH and inhibitors upon the catalytic activity of the dihydroorotase of multienzymatic protein ...
|
|
72692
|
The effects of pH and inhibitors upon the catalytic activity of the dihydroorotase of multienzymatic protein ...
|
|
72693
|
The effects of pH and inhibitors upon the catalytic activity of the dihydroorotase of multienzymatic protein ...
|
|
72694
|
The effects of pH and inhibitors upon the catalytic activity of the dihydroorotase of multienzymatic protein ...
|
|
72695
|
The effects of pH and inhibitors upon the catalytic activity of the dihydroorotase of multienzymatic protein ...
|
|
72696
|
The effects of pH and inhibitors upon the catalytic activity of the dihydroorotase of multienzymatic protein ...
|
|
72697
|
The effects of pH and inhibitors upon the catalytic activity of the dihydroorotase of multienzymatic protein ...
|
|
72698
|
The effects of pH and inhibitors upon the catalytic activity of the dihydroorotase of multienzymatic protein ...
|
|
72699
|
The effects of pH and inhibitors upon the catalytic activity of the dihydroorotase of multienzymatic protein ...
|
|
72700
|
The effects of pH and inhibitors upon the catalytic activity of the dihydroorotase of multienzymatic protein ...
|
|
72701
|
The effects of pH and inhibitors upon the catalytic activity of the dihydroorotase of multienzymatic protein ...
|
|
72702
|
The effects of pH and inhibitors upon the catalytic activity of the dihydroorotase of multienzymatic protein ...
|
|
72703
|
The effects of pH and inhibitors upon the catalytic activity of the dihydroorotase of multienzymatic protein ...
|
|
72704
|
The effects of pH and inhibitors upon the catalytic activity of the dihydroorotase of multienzymatic protein ...
|
|
72705
|
Probing the catalytic mechanism of S-ribosylhomocysteinase (LuxS) with catalytic intermediates and substrate ...
|
|
72706
|
Probing the catalytic mechanism of S-ribosylhomocysteinase (LuxS) with catalytic intermediates and substrate ...
|
|
72707
|
Probing the catalytic mechanism of S-ribosylhomocysteinase (LuxS) with catalytic intermediates and substrate ...
|
|
72708
|
Probing the catalytic mechanism of S-ribosylhomocysteinase (LuxS) with catalytic intermediates and substrate ...
|
|
72709
|
Probing the catalytic mechanism of S-ribosylhomocysteinase (LuxS) with catalytic intermediates and substrate ...
|
|
72710
|
Probing the catalytic mechanism of S-ribosylhomocysteinase (LuxS) with catalytic intermediates and substrate ...
|
|
72711
|
Probing the catalytic mechanism of S-ribosylhomocysteinase (LuxS) with catalytic intermediates and substrate ...
|
|
72712
|
Probing the catalytic mechanism of S-ribosylhomocysteinase (LuxS) with catalytic intermediates and substrate ...
|
|
72713
|
Probing the catalytic mechanism of S-ribosylhomocysteinase (LuxS) with catalytic intermediates and substrate ...
|
|
72714
|
Probing the catalytic mechanism of S-ribosylhomocysteinase (LuxS) with catalytic intermediates and substrate ...
|
|
72715
|
Probing the catalytic mechanism of S-ribosylhomocysteinase (LuxS) with catalytic intermediates and substrate ...
|
|
72716
|
Probing the catalytic mechanism of S-ribosylhomocysteinase (LuxS) with catalytic intermediates and substrate ...
|
|
72717
|
Probing the catalytic mechanism of S-ribosylhomocysteinase (LuxS) with catalytic intermediates and substrate ...
|
|
72718
|
Probing the catalytic mechanism of S-ribosylhomocysteinase (LuxS) with catalytic intermediates and substrate ...
|
|
72719
|
Rat liver aromatic L-amino acid decarboxylase: spectroscopic and kinetic analysis of the coenzyme and reaction ...
|
|
72720
|
Rat liver aromatic L-amino acid decarboxylase: spectroscopic and kinetic analysis of the coenzyme and reaction ...
|
|
72721
|
Rat liver aromatic L-amino acid decarboxylase: spectroscopic and kinetic analysis of the coenzyme and reaction ...
|
|
72722
|
Cloning and sequencing of Escherichia coli ubiC and purification of chorismate lyase
|
|
72723
|
In Vitro interaction between yeast frataxin and superoxide dismutases: Influence of mitochondrial metals
|
|
72724
|
In Vitro interaction between yeast frataxin and superoxide dismutases: Influence of mitochondrial metals
|
|
72725
|
The kinase activity of the antisigma factor SpoIIAB is required for activation as well as inhibition of ...
|
|
72726
|
The kinase activity of the antisigma factor SpoIIAB is required for activation as well as inhibition of ...
|
|
72727
|
Mechanism of Ca2+ activation of the NADPH oxidase 5 (NOX5)
|
|
72728
|
Mechanism of Ca2+ activation of the NADPH oxidase 5 (NOX5)
|
|
72729
|
Mechanism of Ca2+ activation of the NADPH oxidase 5 (NOX5)
|
|
72730
|
Mechanism of Ca2+ activation of the NADPH oxidase 5 (NOX5)
|
|
72731
|
Mechanism of Ca2+ activation of the NADPH oxidase 5 (NOX5)
|
|
72732
|
Identification of monocarboxylate transporter 8 as a specific thyroid hormone transporter
|
|
72733
|
Identification of monocarboxylate transporter 8 as a specific thyroid hormone transporter
|
|
72734
|
Identification of monocarboxylate transporter 8 as a specific thyroid hormone transporter
|
|
72735
|
Identification and characterization of a novel Drosophila 3'-phosphoadenosine 5'-phosphosulfate transporter.
|
|
72736
|
Transport of cytokinins mediated by purine transporters of the PUP family expressed in phloem, hydathodes, and ...
|
|
72737
|
Transport of cytokinins mediated by purine transporters of the PUP family expressed in phloem, hydathodes, and ...
|
|
72738
|
Formation of functional cross- species heterodimers of ornithine decarboxylase
|
|
72739
|
Formation of functional cross- species heterodimers of ornithine decarboxylase
|
|
72740
|
Formation of functional cross- species heterodimers of ornithine decarboxylase
|
|
72741
|
Formation of functional cross- species heterodimers of ornithine decarboxylase
|
|
72742
|
Formation of functional cross- species heterodimers of ornithine decarboxylase
|
|
72743
|
Formation of functional cross- species heterodimers of ornithine decarboxylase
|
|
72744
|
Formation of functional cross- species heterodimers of ornithine decarboxylase
|
|
72745
|
Formation of functional cross- species heterodimers of ornithine decarboxylase
|
|
72746
|
Formation of functional cross- species heterodimers of ornithine decarboxylase
|
|
72747
|
Formation of functional cross- species heterodimers of ornithine decarboxylase
|
|
72748
|
Formation of functional cross- species heterodimers of ornithine decarboxylase
|
|
72749
|
Formation of functional cross- species heterodimers of ornithine decarboxylase
|
|
72750
|
Formation of functional cross- species heterodimers of ornithine decarboxylase
|
|
72751
|
Formation of functional cross- species heterodimers of ornithine decarboxylase
|
|
72752
|
Formation of functional cross- species heterodimers of ornithine decarboxylase
|
|
72753
|
Formation of functional cross- species heterodimers of ornithine decarboxylase
|
|
72754
|
Formation of functional cross- species heterodimers of ornithine decarboxylase
|
|
72755
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72756
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72757
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72758
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72759
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72760
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72761
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72762
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72763
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72764
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72765
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72766
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72767
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72768
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72769
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72770
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72771
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72772
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72773
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72774
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72775
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72776
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72777
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72778
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72779
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72780
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72781
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72782
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72783
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72784
|
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha- barrel fold basic ...
|
|
72785
|
D-amino acid N-acetyltransferase of Saccharomyces cerevisiae: a close homologue of histone acetyltransferase ...
|
|
72786
|
D-amino acid N-acetyltransferase of Saccharomyces cerevisiae: a close homologue of histone acetyltransferase ...
|
|
72787
|
Acetyltransfer precedes uridylyltransfer in the formation of UDP-N-acetylglucosamine in separable active sites ...
|
|
72788
|
Acetyltransfer precedes uridylyltransfer in the formation of UDP-N-acetylglucosamine in separable active sites ...
|
|
72789
|
Acetyltransfer precedes uridylyltransfer in the formation of UDP-N-acetylglucosamine in separable active sites ...
|
|
72790
|
Acetyltransfer precedes uridylyltransfer in the formation of UDP-N-acetylglucosamine in separable active sites ...
|
|
72791
|
Acetyltransfer precedes uridylyltransfer in the formation of UDP-N-acetylglucosamine in separable active sites ...
|
|
72792
|
Acetyltransfer precedes uridylyltransfer in the formation of UDP-N-acetylglucosamine in separable active sites ...
|
|
72793
|
Acetyltransfer precedes uridylyltransfer in the formation of UDP-N-acetylglucosamine in separable active sites ...
|
|
72794
|
Acetyltransfer precedes uridylyltransfer in the formation of UDP-N-acetylglucosamine in separable active sites ...
|
|
72795
|
Acetyltransfer precedes uridylyltransfer in the formation of UDP-N-acetylglucosamine in separable active sites ...
|
|
72796
|
Acetyltransfer precedes uridylyltransfer in the formation of UDP-N-acetylglucosamine in separable active sites ...
|
|
72797
|
Acetyltransfer precedes uridylyltransfer in the formation of UDP-N-acetylglucosamine in separable active sites ...
|
|
72798
|
Acetyltransfer precedes uridylyltransfer in the formation of UDP-N-acetylglucosamine in separable active sites ...
|
|
72799
|
Acetyltransfer precedes uridylyltransfer in the formation of UDP-N-acetylglucosamine in separable active sites ...
|
|
72800
|
Acetyltransfer precedes uridylyltransfer in the formation of UDP-N-acetylglucosamine in separable active sites ...
|
|
72801
|
Acetyltransfer precedes uridylyltransfer in the formation of UDP-N-acetylglucosamine in separable active sites ...
|
|
72802
|
Crystal structure of the 2-oxoglutarate- and Fe(II)-dependent lysyl hydroxylase JMJD6
|
|
72803
|
Overexpression in Escherichia coli of soluble aristolochene synthase from Penicillium roqueforti.
|
|
72804
|
Aristolochene synthase: purification, molecular cloning, high-level expression in Escherichia coli, and ...
|
|
72805
|
Aristolochene synthase: purification, molecular cloning, high-level expression in Escherichia coli, and ...
|
|
72806
|
Aristolochene synthase: purification, molecular cloning, high-level expression in Escherichia coli, and ...
|
|
72807
|
Reaction of dopa decarboxylase with L-aromatic amino acids under aerobic and anaerobic conditions
|
|
72808
|
Reaction of dopa decarboxylase with L-aromatic amino acids under aerobic and anaerobic conditions
|
|
72809
|
Reaction of dopa decarboxylase with L-aromatic amino acids under aerobic and anaerobic conditions
|
|
72810
|
Reaction of dopa decarboxylase with L-aromatic amino acids under aerobic and anaerobic conditions
|
|
72811
|
Reaction of dopa decarboxylase with L-aromatic amino acids under aerobic and anaerobic conditions
|
|
72812
|
Reaction of dopa decarboxylase with L-aromatic amino acids under aerobic and anaerobic conditions
|
|
72813
|
Reaction of dopa decarboxylase with L-aromatic amino acids under aerobic and anaerobic conditions
|
|
72814
|
Reaction of dopa decarboxylase with L-aromatic amino acids under aerobic and anaerobic conditions
|
|
72815
|
Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute ...
|
|
72816
|
Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute ...
|
|
72817
|
Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute ...
|
|
72818
|
Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute ...
|
|
72819
|
Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute ...
|
|
72820
|
Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute ...
|
|
72821
|
Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute ...
|
|
72822
|
Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute ...
|
|
72823
|
Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute ...
|
|
72824
|
Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute ...
|
|
72825
|
Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute ...
|
|
72826
|
Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute ...
|
|
72827
|
Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute ...
|
|
72828
|
Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute ...
|
|
72829
|
Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute ...
|
|
72830
|
Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute ...
|
|
72831
|
Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute ...
|
|
72832
|
Formation of 4-hydroxybenzoate in Escherichia coli: characterization of the ubiC gene and its encoded enzyme ...
|
|
72833
|
Remdesivir Is Effective in Combating COVID-19 because It Is a Better Substrate than ATP for the Viral ...
|
|
72834
|
Remdesivir Is Effective in Combating COVID-19 because It Is a Better Substrate than ATP for the Viral ...
|
|
72835
|
Remdesivir Is Effective in Combating COVID-19 because It Is a Better Substrate than ATP for the Viral ...
|
|
72836
|
Remdesivir Is Effective in Combating COVID-19 because It Is a Better Substrate than ATP for the Viral ...
|
|
72837
|
Remdesivir Is Effective in Combating COVID-19 because It Is a Better Substrate than ATP for the Viral ...
|
|
72838
|
Remdesivir Is Effective in Combating COVID-19 because It Is a Better Substrate than ATP for the Viral ...
|
|
72839
|
Remdesivir Is Effective in Combating COVID-19 because It Is a Better Substrate than ATP for the Viral ...
|
|
72840
|
Remdesivir Is Effective in Combating COVID-19 because It Is a Better Substrate than ATP for the Viral ...
|
|
72841
|
Remdesivir Is Effective in Combating COVID-19 because It Is a Better Substrate than ATP for the Viral ...
|
|
72842
|
Isocitrate lyase from Phycomyces blakesleeanus. The role of Mg2+ ions, kinetics and evidence for two classes ...
|
|
72843
|
Isocitrate lyase from Phycomyces blakesleeanus. The role of Mg2+ ions, kinetics and evidence for two classes ...
|
|
72844
|
Isocitrate lyase from Phycomyces blakesleeanus. The role of Mg2+ ions, kinetics and evidence for two classes ...
|
|
72845
|
Isocitrate lyase from Phycomyces blakesleeanus. The role of Mg2+ ions, kinetics and evidence for two classes ...
|
|
72846
|
Isocitrate lyase from Phycomyces blakesleeanus. The role of Mg2+ ions, kinetics and evidence for two classes ...
|
|
72847
|
Isocitrate lyase from Phycomyces blakesleeanus. The role of Mg2+ ions, kinetics and evidence for two classes ...
|
|
72848
|
Isocitrate lyase from Phycomyces blakesleeanus. The role of Mg2+ ions, kinetics and evidence for two classes ...
|
|
72849
|
Isocitrate lyase from Phycomyces blakesleeanus. The role of Mg2+ ions, kinetics and evidence for two classes ...
|
|
72850
|
Isocitrate lyase from Phycomyces blakesleeanus. The role of Mg2+ ions, kinetics and evidence for two classes ...
|
|
72851
|
Isocitrate lyase from Phycomyces blakesleeanus. The role of Mg2+ ions, kinetics and evidence for two classes ...
|
|
72852
|
Isocitrate lyase from Phycomyces blakesleeanus. The role of Mg2+ ions, kinetics and evidence for two classes ...
|
|
72853
|
Isocitrate lyase from Phycomyces blakesleeanus. The role of Mg2+ ions, kinetics and evidence for two classes ...
|
|
72854
|
Isocitrate lyase from Phycomyces blakesleeanus. The role of Mg2+ ions, kinetics and evidence for two classes ...
|
|
72855
|
Isocitrate lyase from Phycomyces blakesleeanus. The role of Mg2+ ions, kinetics and evidence for two classes ...
|
|
72856
|
Isocitrate lyase from Phycomyces blakesleeanus. The role of Mg2+ ions, kinetics and evidence for two classes ...
|
|
72857
|
Isocitrate lyase from Phycomyces blakesleeanus. The role of Mg2+ ions, kinetics and evidence for two classes ...
|
|
72858
|
Isocitrate lyase from Phycomyces blakesleeanus. The role of Mg2+ ions, kinetics and evidence for two classes ...
|
|
72859
|
Acidic residues important for substrate binding and cofactor reactivity in eukaryotic ornithine decarboxylase ...
|
|
72860
|
Acidic residues important for substrate binding and cofactor reactivity in eukaryotic ornithine decarboxylase ...
|
|
72861
|
Acidic residues important for substrate binding and cofactor reactivity in eukaryotic ornithine decarboxylase ...
|
|
72862
|
Acidic residues important for substrate binding and cofactor reactivity in eukaryotic ornithine decarboxylase ...
|
|
72863
|
Acidic residues important for substrate binding and cofactor reactivity in eukaryotic ornithine decarboxylase ...
|
|
72864
|
Acidic residues important for substrate binding and cofactor reactivity in eukaryotic ornithine decarboxylase ...
|
|
72865
|
Acidic residues important for substrate binding and cofactor reactivity in eukaryotic ornithine decarboxylase ...
|
|
72866
|
Acidic residues important for substrate binding and cofactor reactivity in eukaryotic ornithine decarboxylase ...
|
|
72867
|
Acidic residues important for substrate binding and cofactor reactivity in eukaryotic ornithine decarboxylase ...
|
|
72868
|
Acidic residues important for substrate binding and cofactor reactivity in eukaryotic ornithine decarboxylase ...
|
|
72869
|
Acidic residues important for substrate binding and cofactor reactivity in eukaryotic ornithine decarboxylase ...
|
|
72870
|
Acidic residues important for substrate binding and cofactor reactivity in eukaryotic ornithine decarboxylase ...
|
|
72871
|
Acidic residues important for substrate binding and cofactor reactivity in eukaryotic ornithine decarboxylase ...
|
|
72872
|
Acidic residues important for substrate binding and cofactor reactivity in eukaryotic ornithine decarboxylase ...
|
|
72873
|
Acidic residues important for substrate binding and cofactor reactivity in eukaryotic ornithine decarboxylase ...
|
|
72874
|
Acidic residues important for substrate binding and cofactor reactivity in eukaryotic ornithine decarboxylase ...
|
|
72875
|
Cloning, purification and characterisation of cytosolic fructose-1,6-bisphosphatase from mung bean (Vigna ...
|
|
72876
|
Cloning, purification and characterisation of cytosolic fructose-1,6-bisphosphatase from mung bean (Vigna ...
|
|
72877
|
Cloning, purification and characterisation of cytosolic fructose-1,6-bisphosphatase from mung bean (Vigna ...
|
|
72878
|
Cloning, purification and characterisation of cytosolic fructose-1,6-bisphosphatase from mung bean (Vigna ...
|
|
72879
|
Cloning, purification and characterisation of cytosolic fructose-1,6-bisphosphatase from mung bean (Vigna ...
|
|
72880
|
Cloning, purification and characterisation of cytosolic fructose-1,6-bisphosphatase from mung bean (Vigna ...
|
|
72881
|
Cloning, purification and characterisation of cytosolic fructose-1,6-bisphosphatase from mung bean (Vigna ...
|
|
72882
|
Cloning, purification and characterisation of cytosolic fructose-1,6-bisphosphatase from mung bean (Vigna ...
|
|
72883
|
Cloning, purification and characterisation of cytosolic fructose-1,6-bisphosphatase from mung bean (Vigna ...
|
|
72884
|
Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in ...
|
|
72885
|
The AtProT family. Compatible solute transporters with similar substrate specificity but differential ...
|
|
72886
|
The AtProT family. Compatible solute transporters with similar substrate specificity but differential ...
|
|
72887
|
The AtProT family. Compatible solute transporters with similar substrate specificity but differential ...
|
|
72888
|
The AtProT family. Compatible solute transporters with similar substrate specificity but differential ...
|
|
72889
|
The AtProT family. Compatible solute transporters with similar substrate specificity but differential ...
|
|
72890
|
The AtProT family. Compatible solute transporters with similar substrate specificity but differential ...
|
|
72891
|
The AtProT family. Compatible solute transporters with similar substrate specificity but differential ...
|
|
72892
|
The AtProT family. Compatible solute transporters with similar substrate specificity but differential ...
|
|
72893
|
The AtProT family. Compatible solute transporters with similar substrate specificity but differential ...
|
|
72894
|
Purification and kinetic studies of recombinant gibberellin dioxygenases
|
|
72895
|
Purification and kinetic studies of recombinant gibberellin dioxygenases
|
|
72896
|
Methylenetetrahydrofolate reductase from Escherichia coli: elucidation of the kinetic mechanism by ...
|
|
72897
|
Methylenetetrahydrofolate reductase from Escherichia coli: elucidation of the kinetic mechanism by ...
|
|
72898
|
Methylenetetrahydrofolate reductase from Escherichia coli: elucidation of the kinetic mechanism by ...
|
|
72899
|
Methylenetetrahydrofolate reductase from Escherichia coli: elucidation of the kinetic mechanism by ...
|
|
72900
|
Methylenetetrahydrofolate reductase from Escherichia coli: elucidation of the kinetic mechanism by ...
|
|
72901
|
Methylenetetrahydrofolate reductase from Escherichia coli: elucidation of the kinetic mechanism by ...
|
|
72902
|
Methylenetetrahydrofolate reductase from Escherichia coli: elucidation of the kinetic mechanism by ...
|
|
72903
|
Methylenetetrahydrofolate reductase from Escherichia coli: elucidation of the kinetic mechanism by ...
|
|
72904
|
Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. ...
|
|
72905
|
Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. ...
|
|
72906
|
Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. ...
|
|
72907
|
Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. ...
|
|
72908
|
Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. ...
|
|
72909
|
Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. ...
|
|
72910
|
Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. ...
|
|
72911
|
Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. ...
|
|
72912
|
Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. ...
|
|
72913
|
Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. ...
|
|
72914
|
Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. ...
|
|
72915
|
Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. ...
|
|
72916
|
Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. ...
|
|
72917
|
Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. ...
|
|
72918
|
Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. ...
|
|
72919
|
Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. ...
|
|
72920
|
Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. ...
|
|
72921
|
Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. ...
|
|
72922
|
Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-Zeatin
|
|
72923
|
Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-Zeatin
|
|
72924
|
Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-Zeatin
|
|
72925
|
Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-Zeatin
|
|
72926
|
Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-Zeatin
|
|
72927
|
Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-Zeatin
|
|
72928
|
Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 ...
|
|
72929
|
Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 ...
|
|
72930
|
Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 ...
|
|
72931
|
Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 ...
|
|
72932
|
Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 ...
|
|
72933
|
Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 ...
|
|
72934
|
Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 ...
|
|
72935
|
Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 ...
|
|
72936
|
Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 ...
|
|
72937
|
Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 ...
|
|
72938
|
Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 ...
|
|
72939
|
Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 ...
|
|
72940
|
Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 ...
|
|
72941
|
Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 ...
|
|
72942
|
Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 ...
|
|
72943
|
Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 ...
|
|
72944
|
Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 ...
|
|
72945
|
Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 ...
|
|
72946
|
Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 ...
|
|
72947
|
Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 ...
|
|
72948
|
Mutation of Asn28 disrupts the dimerization and enzymatic activity of SARS 3CL(pro)
|
|
72949
|
Mutation of Asn28 disrupts the dimerization and enzymatic activity of SARS 3CL(pro)
|
|
72950
|
A quantitative method for the specific assessment of caspase-6 activity in cell culture
|
|
72951
|
A quantitative method for the specific assessment of caspase-6 activity in cell culture
|
|
72952
|
A quantitative method for the specific assessment of caspase-6 activity in cell culture
|
|
72953
|
A quantitative method for the specific assessment of caspase-6 activity in cell culture
|
|
72954
|
Characterization of a hormone-inducible, high affinity adenosine 3'-5'-cyclic monophosphate phosphodiesterase ...
|
|
72955
|
Characterization of a hormone-inducible, high affinity adenosine 3'-5'-cyclic monophosphate phosphodiesterase ...
|
|
72956
|
Characterization of a hormone-inducible, high affinity adenosine 3'-5'-cyclic monophosphate phosphodiesterase ...
|
|
72957
|
Characterization of a hormone-inducible, high affinity adenosine 3'-5'-cyclic monophosphate phosphodiesterase ...
|
|
72958
|
Sesquiterpene lactones are potent and irreversible inhibitors of the antibacterial target enzyme MurA
|
|
72959
|
Sesquiterpene lactones are potent and irreversible inhibitors of the antibacterial target enzyme MurA
|
|
72960
|
Sesquiterpene lactones are potent and irreversible inhibitors of the antibacterial target enzyme MurA
|
|
72961
|
Sesquiterpene lactones are potent and irreversible inhibitors of the antibacterial target enzyme MurA
|
|
72962
|
Sesquiterpene lactones are potent and irreversible inhibitors of the antibacterial target enzyme MurA
|
|
72963
|
Sesquiterpene lactones are potent and irreversible inhibitors of the antibacterial target enzyme MurA
|
|
72964
|
Sesquiterpene lactones are potent and irreversible inhibitors of the antibacterial target enzyme MurA
|
|
72965
|
Sesquiterpene lactones are potent and irreversible inhibitors of the antibacterial target enzyme MurA
|
|
72966
|
Sesquiterpene lactones are potent and irreversible inhibitors of the antibacterial target enzyme MurA
|
|
72967
|
Development of a whole- cell assay for peptidoglycan biosynthesis inhibitors
|
|
72968
|
The nature of Staphylococcus aureus MurA and MurZ and approaches for detection of peptidoglycan biosynthesis ...
|
|
72969
|
The nature of Staphylococcus aureus MurA and MurZ and approaches for detection of peptidoglycan biosynthesis ...
|
|
72970
|
Identification and characterization of new inhibitors of the Escherichia coli MurA enzyme
|
|
72971
|
Identification and characterization of new inhibitors of the Escherichia coli MurA enzyme
|
|
72972
|
Identification and characterization of new inhibitors of the Escherichia coli MurA enzyme
|
|
72973
|
Identification and characterization of new inhibitors of the Escherichia coli MurA enzyme
|
|
72974
|
Identification and characterization of new inhibitors of the Escherichia coli MurA enzyme
|
|
72975
|
Identification and characterization of new inhibitors of the Escherichia coli MurA enzyme
|
|
72976
|
Identification and characterization of new inhibitors of the Escherichia coli MurA enzyme
|
|
72977
|
Identification and characterization of new inhibitors of the Escherichia coli MurA enzyme
|
|
72978
|
Identification and characterization of new inhibitors of the Escherichia coli MurA enzyme
|
|
72979
|
Identification and characterization of new inhibitors of the Escherichia coli MurA enzyme
|
|
72980
|
Identification and characterization of new inhibitors of the Escherichia coli MurA enzyme
|
|
72981
|
Identification and characterization of new inhibitors of the Escherichia coli MurA enzyme
|
|
72982
|
Identification and characterization of new inhibitors of the Escherichia coli MurA enzyme
|
|
72983
|
Identification and characterization of new inhibitors of the Escherichia coli MurA enzyme
|
|
72984
|
2-Aminotetralones: novel inhibitors of MurA and MurZ
|
|
72985
|
2-Aminotetralones: novel inhibitors of MurA and MurZ
|
|
72986
|
2-Aminotetralones: novel inhibitors of MurA and MurZ
|
|
72987
|
2-Aminotetralones: novel inhibitors of MurA and MurZ
|
|
72988
|
2-Aminotetralones: novel inhibitors of MurA and MurZ
|
|
72989
|
2-Aminotetralones: novel inhibitors of MurA and MurZ
|
|
72990
|
2-Aminotetralones: novel inhibitors of MurA and MurZ
|
|
72991
|
2-Aminotetralones: novel inhibitors of MurA and MurZ
|
|
72992
|
2-Aminotetralones: novel inhibitors of MurA and MurZ
|
|
72993
|
2-Aminotetralones: novel inhibitors of MurA and MurZ
|
|
72994
|
2-Aminotetralones: novel inhibitors of MurA and MurZ
|
|
72995
|
2-Aminotetralones: novel inhibitors of MurA and MurZ
|
|
72996
|
2-Aminotetralones: novel inhibitors of MurA and MurZ
|
|
72997
|
2-Aminotetralones: novel inhibitors of MurA and MurZ
|
|
72998
|
2-Aminotetralones: novel inhibitors of MurA and MurZ
|
|
72999
|
A second isoform of 3-ketoacyl-CoA thiolase found in Caenorhabditis elegans, which is similar to sterol ...
|
|
73000
|
Molecular cloning and characterization of a cytochrome P450 in sanguinarine biosynthesis from Eschscholzia ...
|





 If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.
If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.