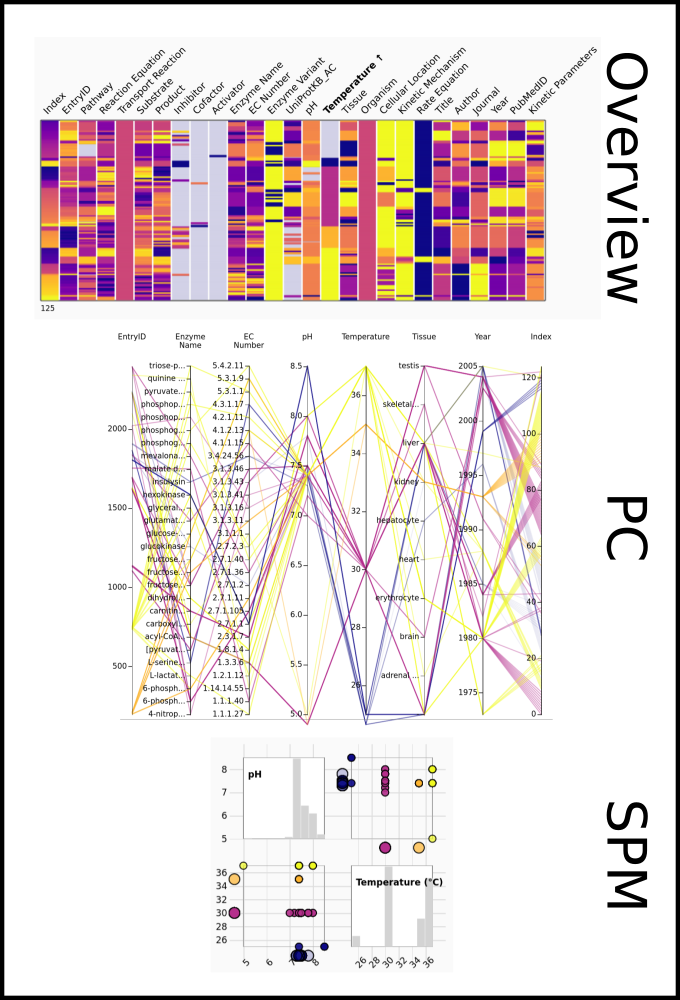
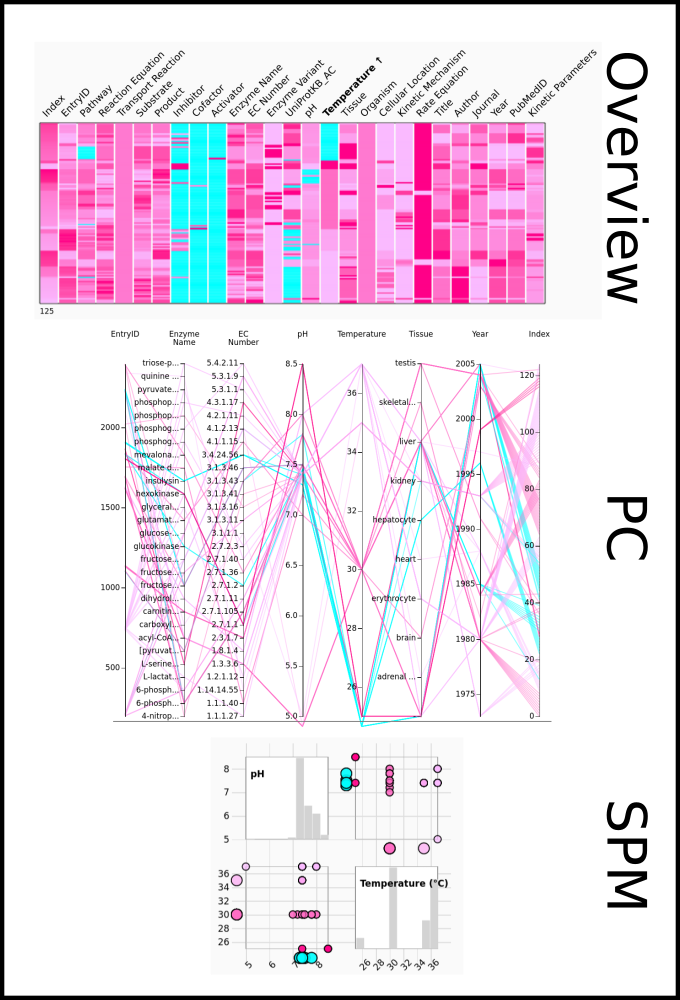
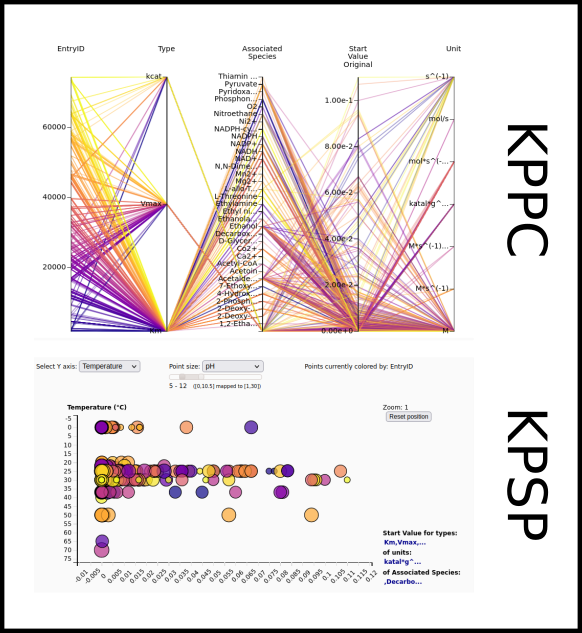
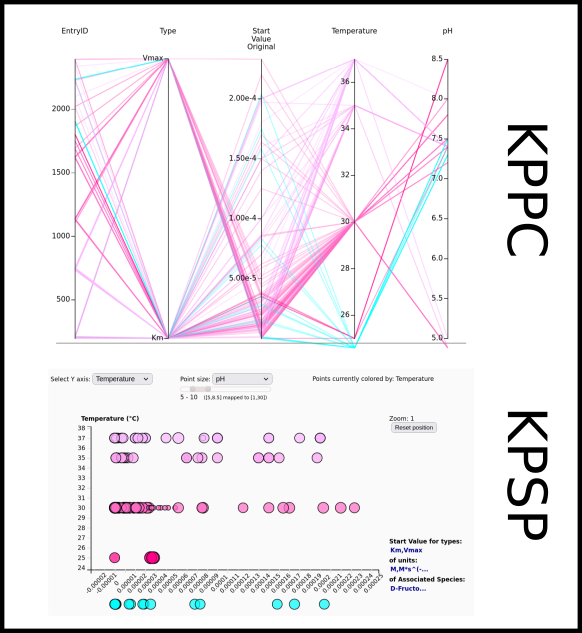
To improve SABIO-RK data interoperability, semantic markup was added to web pages as described and defined by
This structured information makes it easier to discover, collate and analyse our data.
|
71001
|
The Hepatitis E virus intraviral interactome
|
|
71002
|
The Hepatitis E virus intraviral interactome
|
|
71003
|
The Hepatitis E virus intraviral interactome
|
|
71004
|
The Hepatitis E virus intraviral interactome
|
|
71005
|
The Hepatitis E virus intraviral interactome
|
|
71006
|
The Hepatitis E virus intraviral interactome
|
|
71007
|
The Hepatitis E virus intraviral interactome
|
|
71008
|
The Hepatitis E virus intraviral interactome
|
|
71009
|
The Hepatitis E virus intraviral interactome
|
|
71010
|
The Hepatitis E virus intraviral interactome
|
|
71011
|
The octahaem MccA is a haem c-copper sulfite reductase.
|
|
71012
|
The octahaem MccA is a haem c-copper sulfite reductase.
|
|
71013
|
The octahaem MccA is a haem c-copper sulfite reductase.
|
|
71014
|
The octahaem MccA is a haem c-copper sulfite reductase.
|
|
71015
|
Purification and mutagenesis of LpxL, the lauroyltransferase of Escherichia coli lipid A biosynthesis.
|
|
71016
|
Purification and mutagenesis of LpxL, the lauroyltransferase of Escherichia coli lipid A biosynthesis.
|
|
71017
|
Structure, functional characterization, and evolution of the dihydroorotase domain of human CAD
|
|
71018
|
Structure, functional characterization, and evolution of the dihydroorotase domain of human CAD
|
|
71019
|
Structural and functional analyses of human tryptophan 2,3-dioxygenase
|
|
71020
|
Structural and functional analyses of human tryptophan 2,3-dioxygenase
|
|
71021
|
Structural and functional analyses of human tryptophan 2,3-dioxygenase
|
|
71022
|
Structural and functional analyses of human tryptophan 2,3-dioxygenase
|
|
71023
|
Structural and functional analyses of human tryptophan 2,3-dioxygenase
|
|
71024
|
Structural and functional analyses of human tryptophan 2,3-dioxygenase
|
|
71025
|
Structural and functional analyses of human tryptophan 2,3-dioxygenase
|
|
71026
|
Catalytic activity of human indoleamine 2,3-dioxygenase (hIDO1) at low oxygen
|
|
71027
|
Catalytic activity of human indoleamine 2,3-dioxygenase (hIDO1) at low oxygen
|
|
71028
|
Catalytic activity of human indoleamine 2,3-dioxygenase (hIDO1) at low oxygen
|
|
71029
|
Catalytic activity of human indoleamine 2,3-dioxygenase (hIDO1) at low oxygen
|
|
71030
|
Catalytic activity of human indoleamine 2,3-dioxygenase (hIDO1) at low oxygen
|
|
71031
|
Catalytic activity of human indoleamine 2,3-dioxygenase (hIDO1) at low oxygen
|
|
71032
|
Catalytic activity of human indoleamine 2,3-dioxygenase (hIDO1) at low oxygen
|
|
71033
|
Catalytic activity of human indoleamine 2,3-dioxygenase (hIDO1) at low oxygen
|
|
71034
|
Catalytic activity of human indoleamine 2,3-dioxygenase (hIDO1) at low oxygen
|
|
71035
|
Catalytic activity of human indoleamine 2,3-dioxygenase (hIDO1) at low oxygen
|
|
71036
|
Catalytic activity of human indoleamine 2,3-dioxygenase (hIDO1) at low oxygen
|
|
71037
|
Catalytic activity of human indoleamine 2,3-dioxygenase (hIDO1) at low oxygen
|
|
71038
|
Catalytic activity of human indoleamine 2,3-dioxygenase (hIDO1) at low oxygen
|
|
71039
|
Catalytic activity of human indoleamine 2,3-dioxygenase (hIDO1) at low oxygen
|
|
71040
|
Catalytic activity of human indoleamine 2,3-dioxygenase (hIDO1) at low oxygen
|
|
71041
|
Catalytic activity of human indoleamine 2,3-dioxygenase (hIDO1) at low oxygen
|
|
71042
|
Catalytic activity of human indoleamine 2,3-dioxygenase (hIDO1) at low oxygen
|
|
71043
|
Catalytic activity of human indoleamine 2,3-dioxygenase (hIDO1) at low oxygen
|
|
71044
|
Catalytic activity of human indoleamine 2,3-dioxygenase (hIDO1) at low oxygen
|
|
71045
|
Catalytic activity of human indoleamine 2,3-dioxygenase (hIDO1) at low oxygen
|
|
71046
|
Human indolylamine 2,3-dioxygenase. Its tissue distribution, and characterization of the placental enzyme
|
|
71047
|
Human indolylamine 2,3-dioxygenase. Its tissue distribution, and characterization of the placental enzyme
|
|
71048
|
Human indolylamine 2,3-dioxygenase. Its tissue distribution, and characterization of the placental enzyme
|
|
71049
|
Reassessment of the reaction mechanism in the heme dioxygenases
|
|
71050
|
Reassessment of the reaction mechanism in the heme dioxygenases
|
|
71051
|
Reassessment of the reaction mechanism in the heme dioxygenases
|
|
71052
|
Reassessment of the reaction mechanism in the heme dioxygenases
|
|
71053
|
Reassessment of the reaction mechanism in the heme dioxygenases
|
|
71054
|
Reassessment of the reaction mechanism in the heme dioxygenases
|
|
71055
|
Reassessment of the reaction mechanism in the heme dioxygenases
|
|
71056
|
Reassessment of the reaction mechanism in the heme dioxygenases
|
|
71057
|
Reassessment of the reaction mechanism in the heme dioxygenases
|
|
71058
|
Biochemical characterization of the very long-chain fatty acid elongase ELOVL7.
|
|
71059
|
Biochemical characterization of the very long-chain fatty acid elongase ELOVL7.
|
|
71060
|
Fatty acid elongation in yeast--biochemical characteristics of the enzyme system and isolation of ...
|
|
71061
|
Fatty acid elongation in yeast--biochemical characteristics of the enzyme system and isolation of ...
|
|
71062
|
Fatty acid elongation in yeast--biochemical characteristics of the enzyme system and isolation of ...
|
|
71063
|
Fatty acid elongation in yeast--biochemical characteristics of the enzyme system and isolation of ...
|
|
71064
|
Fatty acid elongation in yeast--biochemical characteristics of the enzyme system and isolation of ...
|
|
71065
|
Fatty acid elongation in yeast--biochemical characteristics of the enzyme system and isolation of ...
|
|
71066
|
PqsD is responsible for the synthesis of 2,4-dihydroxyquinoline, an extracellular metabolite produced by ...
|
|
71067
|
PqsD is responsible for the synthesis of 2,4-dihydroxyquinoline, an extracellular metabolite produced by ...
|
|
71068
|
PqsD is responsible for the synthesis of 2,4-dihydroxyquinoline, an extracellular metabolite produced by ...
|
|
71069
|
The Escherichia coli CysZ is a pH dependent sulfate transporter that can be inhibited by sulfite.
|
|
71070
|
The Escherichia coli CysZ is a pH dependent sulfate transporter that can be inhibited by sulfite.
|
|
71071
|
The Escherichia coli CysZ is a pH dependent sulfate transporter that can be inhibited by sulfite.
|
|
71072
|
The Escherichia coli CysZ is a pH dependent sulfate transporter that can be inhibited by sulfite.
|
|
71073
|
The Escherichia coli CysZ is a pH dependent sulfate transporter that can be inhibited by sulfite.
|
|
71074
|
The Escherichia coli CysZ is a pH dependent sulfate transporter that can be inhibited by sulfite.
|
|
71075
|
The Escherichia coli CysZ is a pH dependent sulfate transporter that can be inhibited by sulfite.
|
|
71076
|
The Escherichia coli CysZ is a pH dependent sulfate transporter that can be inhibited by sulfite.
|
|
71077
|
Structural basis for the catalytic mechanism of a proficient enzyme: orotidine 5'-monophosphate decarboxylase.
|
|
71078
|
Cytidine 5'-triphosphate-dependent biosynthesis of isoprenoids: YgbP protein of Escherichia coli catalyzes the ...
|
|
71079
|
Evidence of a functional requirement for a carbamoylated lysine residue in MurD, MurE and MurF synthetases as ...
|
|
71080
|
Evidence of a functional requirement for a carbamoylated lysine residue in MurD, MurE and MurF synthetases as ...
|
|
71081
|
Evidence of a functional requirement for a carbamoylated lysine residue in MurD, MurE and MurF synthetases as ...
|
|
71082
|
Evidence of a functional requirement for a carbamoylated lysine residue in MurD, MurE and MurF synthetases as ...
|
|
71083
|
Evidence of a functional requirement for a carbamoylated lysine residue in MurD, MurE and MurF synthetases as ...
|
|
71084
|
Evidence of a functional requirement for a carbamoylated lysine residue in MurD, MurE and MurF synthetases as ...
|
|
71085
|
Evidence of a functional requirement for a carbamoylated lysine residue in MurD, MurE and MurF synthetases as ...
|
|
71086
|
Evidence of a functional requirement for a carbamoylated lysine residue in MurD, MurE and MurF synthetases as ...
|
|
71087
|
Evidence of a functional requirement for a carbamoylated lysine residue in MurD, MurE and MurF synthetases as ...
|
|
71088
|
Evidence of a functional requirement for a carbamoylated lysine residue in MurD, MurE and MurF synthetases as ...
|
|
71089
|
Evidence of a functional requirement for a carbamoylated lysine residue in MurD, MurE and MurF synthetases as ...
|
|
71090
|
Evidence of a functional requirement for a carbamoylated lysine residue in MurD, MurE and MurF synthetases as ...
|
|
71091
|
Evidence of a functional requirement for a carbamoylated lysine residue in MurD, MurE and MurF synthetases as ...
|
|
71092
|
Evidence of a functional requirement for a carbamoylated lysine residue in MurD, MurE and MurF synthetases as ...
|
|
71093
|
Evidence of a functional requirement for a carbamoylated lysine residue in MurD, MurE and MurF synthetases as ...
|
|
71094
|
Evidence of a functional requirement for a carbamoylated lysine residue in MurD, MurE and MurF synthetases as ...
|
|
71095
|
Evidence of a functional requirement for a carbamoylated lysine residue in MurD, MurE and MurF synthetases as ...
|
|
71096
|
Evidence of a functional requirement for a carbamoylated lysine residue in MurD, MurE and MurF synthetases as ...
|
|
71097
|
Evidence of a functional requirement for a carbamoylated lysine residue in MurD, MurE and MurF synthetases as ...
|
|
71098
|
Evidence of a functional requirement for a carbamoylated lysine residue in MurD, MurE and MurF synthetases as ...
|
|
71099
|
Evidence of a functional requirement for a carbamoylated lysine residue in MurD, MurE and MurF synthetases as ...
|
|
71100
|
Biosynthesis of riboflavin. Single turnover kinetic analysis of GTP cyclohydrolase II
|
|
71101
|
Biosynthesis of riboflavin. Single turnover kinetic analysis of GTP cyclohydrolase II
|
|
71102
|
Biosynthesis of riboflavin. Single turnover kinetic analysis of GTP cyclohydrolase II
|
|
71103
|
Biosynthesis of riboflavin. Single turnover kinetic analysis of GTP cyclohydrolase II
|
|
71104
|
Molecular conversion of NAD kinase to NADH kinase through single amino acid residue substitution
|
|
71105
|
Molecular conversion of NAD kinase to NADH kinase through single amino acid residue substitution
|
|
71106
|
Molecular conversion of NAD kinase to NADH kinase through single amino acid residue substitution
|
|
71107
|
Molecular conversion of NAD kinase to NADH kinase through single amino acid residue substitution
|
|
71108
|
Molecular conversion of NAD kinase to NADH kinase through single amino acid residue substitution
|
|
71109
|
Molecular conversion of NAD kinase to NADH kinase through single amino acid residue substitution
|
|
71110
|
Molecular conversion of NAD kinase to NADH kinase through single amino acid residue substitution
|
|
71111
|
Molecular conversion of NAD kinase to NADH kinase through single amino acid residue substitution
|
|
71112
|
Molecular conversion of NAD kinase to NADH kinase through single amino acid residue substitution
|
|
71113
|
Molecular conversion of NAD kinase to NADH kinase through single amino acid residue substitution
|
|
71114
|
Molecular conversion of NAD kinase to NADH kinase through single amino acid residue substitution
|
|
71115
|
Molecular conversion of NAD kinase to NADH kinase through single amino acid residue substitution
|
|
71116
|
Molecular conversion of NAD kinase to NADH kinase through single amino acid residue substitution
|
|
71117
|
Molecular conversion of NAD kinase to NADH kinase through single amino acid residue substitution
|
|
71118
|
Molecular conversion of NAD kinase to NADH kinase through single amino acid residue substitution
|
|
71119
|
Molecular conversion of NAD kinase to NADH kinase through single amino acid residue substitution
|
|
71120
|
Molecular conversion of NAD kinase to NADH kinase through single amino acid residue substitution
|
|
71121
|
Molecular conversion of NAD kinase to NADH kinase through single amino acid residue substitution
|
|
71122
|
Molecular conversion of NAD kinase to NADH kinase through single amino acid residue substitution
|
|
71123
|
Molecular conversion of NAD kinase to NADH kinase through single amino acid residue substitution
|
|
71124
|
Molecular conversion of NAD kinase to NADH kinase through single amino acid residue substitution
|
|
71125
|
Molecular conversion of NAD kinase to NADH kinase through single amino acid residue substitution
|
|
71126
|
Dual role for the glutamine phosphoribosylpyrophosphate amidotransferase ammonia channel. Interdomain ...
|
|
71127
|
Dual role for the glutamine phosphoribosylpyrophosphate amidotransferase ammonia channel. Interdomain ...
|
|
71128
|
Dual role for the glutamine phosphoribosylpyrophosphate amidotransferase ammonia channel. Interdomain ...
|
|
71129
|
Dual role for the glutamine phosphoribosylpyrophosphate amidotransferase ammonia channel. Interdomain ...
|
|
71130
|
Dual role for the glutamine phosphoribosylpyrophosphate amidotransferase ammonia channel. Interdomain ...
|
|
71131
|
Dual role for the glutamine phosphoribosylpyrophosphate amidotransferase ammonia channel. Interdomain ...
|
|
71132
|
Dual role for the glutamine phosphoribosylpyrophosphate amidotransferase ammonia channel. Interdomain ...
|
|
71133
|
Dual role for the glutamine phosphoribosylpyrophosphate amidotransferase ammonia channel. Interdomain ...
|
|
71134
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71135
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71136
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71137
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71138
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71139
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71140
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71141
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71142
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71143
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71144
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71145
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71146
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71147
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71148
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71149
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71150
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71151
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71152
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71153
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71154
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71155
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71156
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71157
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71158
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71159
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71160
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71161
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71162
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71163
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71164
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71165
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71166
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71167
|
Riboflavin synthase of Escherichia coli. Effect of single amino acid substitutions on reaction rate and ligand ...
|
|
71168
|
Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts ...
|
|
71169
|
Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts ...
|
|
71170
|
Role of the ortholog and paralog amino acid invariants in the active site of the ...
|
|
71171
|
Role of the ortholog and paralog amino acid invariants in the active site of the ...
|
|
71172
|
Role of the ortholog and paralog amino acid invariants in the active site of the ...
|
|
71173
|
Role of the ortholog and paralog amino acid invariants in the active site of the ...
|
|
71174
|
Role of the ortholog and paralog amino acid invariants in the active site of the ...
|
|
71175
|
Role of the ortholog and paralog amino acid invariants in the active site of the ...
|
|
71176
|
Role of the ortholog and paralog amino acid invariants in the active site of the ...
|
|
71177
|
Role of the ortholog and paralog amino acid invariants in the active site of the ...
|
|
71178
|
Role of the ortholog and paralog amino acid invariants in the active site of the ...
|
|
71179
|
Role of the ortholog and paralog amino acid invariants in the active site of the ...
|
|
71180
|
Role of the ortholog and paralog amino acid invariants in the active site of the ...
|
|
71181
|
Role of the ortholog and paralog amino acid invariants in the active site of the ...
|
|
71182
|
Role of the ortholog and paralog amino acid invariants in the active site of the ...
|
|
71183
|
Role of the ortholog and paralog amino acid invariants in the active site of the ...
|
|
71184
|
Role of the ortholog and paralog amino acid invariants in the active site of the ...
|
|
71185
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71186
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71187
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71188
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71189
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71190
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71191
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71192
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71193
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71194
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71195
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71196
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71197
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71198
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71199
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71200
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71201
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71202
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71203
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71204
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71205
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71206
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71207
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71208
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71209
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71210
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71211
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71212
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71213
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71214
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71215
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71216
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71217
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71218
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71219
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71220
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71221
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71222
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71223
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71224
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71225
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71226
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71227
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71228
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71229
|
Indoleamine 2,3-dioxygenase is the anticancer target for a novel series of potent naphthoquinone-based ...
|
|
71230
|
Engineered glycolytic glyceraldehyde-3-phosphate dehydrogenase binds the anti conformation of NAD+ ...
|
|
71231
|
Engineered glycolytic glyceraldehyde-3-phosphate dehydrogenase binds the anti conformation of NAD+ ...
|
|
71232
|
Engineered glycolytic glyceraldehyde-3-phosphate dehydrogenase binds the anti conformation of NAD+ ...
|
|
71233
|
Engineered glycolytic glyceraldehyde-3-phosphate dehydrogenase binds the anti conformation of NAD+ ...
|
|
71234
|
Engineered glycolytic glyceraldehyde-3-phosphate dehydrogenase binds the anti conformation of NAD+ ...
|
|
71235
|
Engineered glycolytic glyceraldehyde-3-phosphate dehydrogenase binds the anti conformation of NAD+ ...
|
|
71236
|
Engineered glycolytic glyceraldehyde-3-phosphate dehydrogenase binds the anti conformation of NAD+ ...
|
|
71237
|
Engineered glycolytic glyceraldehyde-3-phosphate dehydrogenase binds the anti conformation of NAD+ ...
|
|
71238
|
Engineered glycolytic glyceraldehyde-3-phosphate dehydrogenase binds the anti conformation of NAD+ ...
|
|
71239
|
Engineered glycolytic glyceraldehyde-3-phosphate dehydrogenase binds the anti conformation of NAD+ ...
|
|
71240
|
Molecular cloning and expression of a cDNA encoding an olfactory-specific mouse phenol sulphotransferase
|
|
71241
|
Molecular cloning and expression of a cDNA encoding an olfactory-specific mouse phenol sulphotransferase
|
|
71242
|
Molecular cloning and expression of a cDNA encoding an olfactory-specific mouse phenol sulphotransferase
|
|
71243
|
Expression of pig heart mitochondrial NADP-dependent isocitrate dehydrogenase in Escherichia coli
|
|
71244
|
Expression of pig heart mitochondrial NADP-dependent isocitrate dehydrogenase in Escherichia coli
|
|
71245
|
Sequential hydrolysis of the gamma- and beta-phosphate groups of ATP by the ATP diphosphohydrolase from pig ...
|
|
71246
|
Sequential hydrolysis of the gamma- and beta-phosphate groups of ATP by the ATP diphosphohydrolase from pig ...
|
|
71247
|
Sequential hydrolysis of the gamma- and beta-phosphate groups of ATP by the ATP diphosphohydrolase from pig ...
|
|
71248
|
Sequential hydrolysis of the gamma- and beta-phosphate groups of ATP by the ATP diphosphohydrolase from pig ...
|
|
71249
|
Purification and characterization of specific 3-deoxy-D- manno-octulosonate 8-phosphate phosphatase from ...
|
|
71250
|
Purification and characterization of specific 3-deoxy-D- manno-octulosonate 8-phosphate phosphatase from ...
|
|
71251
|
Purification and characterization of specific 3-deoxy-D- manno-octulosonate 8-phosphate phosphatase from ...
|
|
71252
|
Purification and characterization of specific 3-deoxy-D- manno-octulosonate 8-phosphate phosphatase from ...
|
|
71253
|
Kinetic and thermodynamic characterization of novel alkaline lipase from halotolerant Bacillus gibsonii
|
|
71254
|
Kinetic and thermodynamic characterization of novel alkaline lipase from halotolerant Bacillus gibsonii
|
|
71255
|
Kinetic and thermodynamic characterization of novel alkaline lipase from halotolerant Bacillus gibsonii
|
|
71256
|
Substrate specificity of a new laccase from Trametes polyzona WRF03
|
|
71257
|
Substrate specificity of a new laccase from Trametes polyzona WRF03
|
|
71258
|
Substrate specificity of a new laccase from Trametes polyzona WRF03
|
|
71259
|
Substrate specificity of a new laccase from Trametes polyzona WRF03
|
|
71260
|
Substrate specificity of a new laccase from Trametes polyzona WRF03
|
|
71261
|
Substrate specificity of a new laccase from Trametes polyzona WRF03
|
|
71262
|
The role of serine 167 in human indoleamine 2,3-dioxygenase: a comparison with tryptophan 2,3-dioxygenase
|
|
71263
|
The role of serine 167 in human indoleamine 2,3-dioxygenase: a comparison with tryptophan 2,3-dioxygenase
|
|
71264
|
The role of serine 167 in human indoleamine 2,3-dioxygenase: a comparison with tryptophan 2,3-dioxygenase
|
|
71265
|
The role of serine 167 in human indoleamine 2,3-dioxygenase: a comparison with tryptophan 2,3-dioxygenase
|
|
71266
|
The role of serine 167 in human indoleamine 2,3-dioxygenase: a comparison with tryptophan 2,3-dioxygenase
|
|
71267
|
The role of serine 167 in human indoleamine 2,3-dioxygenase: a comparison with tryptophan 2,3-dioxygenase
|
|
71268
|
The role of serine 167 in human indoleamine 2,3-dioxygenase: a comparison with tryptophan 2,3-dioxygenase
|
|
71269
|
The role of serine 167 in human indoleamine 2,3-dioxygenase: a comparison with tryptophan 2,3-dioxygenase
|
|
71270
|
The role of serine 167 in human indoleamine 2,3-dioxygenase: a comparison with tryptophan 2,3-dioxygenase
|
|
71271
|
The role of serine 167 in human indoleamine 2,3-dioxygenase: a comparison with tryptophan 2,3-dioxygenase
|
|
71272
|
The role of serine 167 in human indoleamine 2,3-dioxygenase: a comparison with tryptophan 2,3-dioxygenase
|
|
71273
|
The role of serine 167 in human indoleamine 2,3-dioxygenase: a comparison with tryptophan 2,3-dioxygenase
|
|
71274
|
The role of serine 167 in human indoleamine 2,3-dioxygenase: a comparison with tryptophan 2,3-dioxygenase
|
|
71275
|
The role of serine 167 in human indoleamine 2,3-dioxygenase: a comparison with tryptophan 2,3-dioxygenase
|
|
71276
|
Thermal stability of isocitrate dehydrogenase from Archaeoglobus fulgidus studied by crystal structure ...
|
|
71277
|
Thermal stability of isocitrate dehydrogenase from Archaeoglobus fulgidus studied by crystal structure ...
|
|
71278
|
Thermal stability of isocitrate dehydrogenase from Archaeoglobus fulgidus studied by crystal structure ...
|
|
71279
|
Thermal stability of isocitrate dehydrogenase from Archaeoglobus fulgidus studied by crystal structure ...
|
|
71280
|
Thermal stability of isocitrate dehydrogenase from Archaeoglobus fulgidus studied by crystal structure ...
|
|
71281
|
Thermal stability of isocitrate dehydrogenase from Archaeoglobus fulgidus studied by crystal structure ...
|
|
71282
|
Thermal stability of isocitrate dehydrogenase from Archaeoglobus fulgidus studied by crystal structure ...
|
|
71283
|
Thermal stability of isocitrate dehydrogenase from Archaeoglobus fulgidus studied by crystal structure ...
|
|
71284
|
Mechanism of product chain length determination and the role of a flexible loop in Escherichia coli ...
|
|
71285
|
Mechanism of product chain length determination and the role of a flexible loop in Escherichia coli ...
|
|
71286
|
Mechanism of product chain length determination and the role of a flexible loop in Escherichia coli ...
|
|
71287
|
Mechanism of product chain length determination and the role of a flexible loop in Escherichia coli ...
|
|
71288
|
Mechanism of product chain length determination and the role of a flexible loop in Escherichia coli ...
|
|
71289
|
Mechanism of product chain length determination and the role of a flexible loop in Escherichia coli ...
|
|
71290
|
Mechanism of product chain length determination and the role of a flexible loop in Escherichia coli ...
|
|
71291
|
Mechanism of product chain length determination and the role of a flexible loop in Escherichia coli ...
|
|
71292
|
Mechanism of product chain length determination and the role of a flexible loop in Escherichia coli ...
|
|
71293
|
Mechanism of product chain length determination and the role of a flexible loop in Escherichia coli ...
|
|
71294
|
Mechanism of product chain length determination and the role of a flexible loop in Escherichia coli ...
|
|
71295
|
Mechanism of product chain length determination and the role of a flexible loop in Escherichia coli ...
|
|
71296
|
Dissection of the bifunctional Escherichia coli N-acetylglucosamine-1-phosphate uridyltransferase enzyme into ...
|
|
71297
|
Dissection of the bifunctional Escherichia coli N-acetylglucosamine-1-phosphate uridyltransferase enzyme into ...
|
|
71298
|
Dissection of the bifunctional Escherichia coli N-acetylglucosamine-1-phosphate uridyltransferase enzyme into ...
|
|
71299
|
Dissection of the bifunctional Escherichia coli N-acetylglucosamine-1-phosphate uridyltransferase enzyme into ...
|
|
71300
|
Dissection of the bifunctional Escherichia coli N-acetylglucosamine-1-phosphate uridyltransferase enzyme into ...
|
|
71301
|
Dissection of the bifunctional Escherichia coli N-acetylglucosamine-1-phosphate uridyltransferase enzyme into ...
|
|
71302
|
Dissection of the bifunctional Escherichia coli N-acetylglucosamine-1-phosphate uridyltransferase enzyme into ...
|
|
71303
|
Dissection of the bifunctional Escherichia coli N-acetylglucosamine-1-phosphate uridyltransferase enzyme into ...
|
|
71304
|
Dissection of the bifunctional Escherichia coli N-acetylglucosamine-1-phosphate uridyltransferase enzyme into ...
|
|
71305
|
Dissection of the bifunctional Escherichia coli N-acetylglucosamine-1-phosphate uridyltransferase enzyme into ...
|
|
71306
|
Dissection of the bifunctional Escherichia coli N-acetylglucosamine-1-phosphate uridyltransferase enzyme into ...
|
|
71307
|
Dissection of the bifunctional Escherichia coli N-acetylglucosamine-1-phosphate uridyltransferase enzyme into ...
|
|
71308
|
Dissection of the bifunctional Escherichia coli N-acetylglucosamine-1-phosphate uridyltransferase enzyme into ...
|
|
71309
|
Dissection of the bifunctional Escherichia coli N-acetylglucosamine-1-phosphate uridyltransferase enzyme into ...
|
|
71310
|
Dissection of the bifunctional Escherichia coli N-acetylglucosamine-1-phosphate uridyltransferase enzyme into ...
|
|
71311
|
Dissection of the bifunctional Escherichia coli N-acetylglucosamine-1-phosphate uridyltransferase enzyme into ...
|
|
71312
|
Engineering a Novel Antibody-Peptide Bispecific Fusion Protein Against MERS-CoV
|
|
71313
|
Engineering a Novel Antibody-Peptide Bispecific Fusion Protein Against MERS-CoV
|
|
71314
|
Engineering a Novel Antibody-Peptide Bispecific Fusion Protein Against MERS-CoV
|
|
71315
|
Engineering a Novel Antibody-Peptide Bispecific Fusion Protein Against MERS-CoV
|
|
71316
|
Engineering a Novel Antibody-Peptide Bispecific Fusion Protein Against MERS-CoV
|
|
71317
|
Engineering a Novel Antibody-Peptide Bispecific Fusion Protein Against MERS-CoV
|
|
71318
|
Engineering a Novel Antibody-Peptide Bispecific Fusion Protein Against MERS-CoV
|
|
71319
|
Structural and functional characterization of polyethylene terephthalate hydrolase from Ideonella sakaiensis.
|
|
71320
|
Structural and functional characterization of polyethylene terephthalate hydrolase from Ideonella sakaiensis.
|
|
71321
|
Structural and functional characterization of polyethylene terephthalate hydrolase from Ideonella sakaiensis.
|
|
71322
|
Structural and functional characterization of polyethylene terephthalate hydrolase from Ideonella sakaiensis.
|
|
71323
|
Structural and functional characterization of polyethylene terephthalate hydrolase from Ideonella sakaiensis.
|
|
71324
|
Sequential O-methylation of tricetin by a single gene product in wheat
|
|
71325
|
Sequential O-methylation of tricetin by a single gene product in wheat
|
|
71326
|
Sequential O-methylation of tricetin by a single gene product in wheat
|
|
71327
|
Sequential O-methylation of tricetin by a single gene product in wheat
|
|
71328
|
Identification of isoliquiritigenin as an activator that stimulates the enzymatic production of glycyrrhetinic ...
|
|
71329
|
Identification of isoliquiritigenin as an activator that stimulates the enzymatic production of glycyrrhetinic ...
|
|
71330
|
A bacterium that degrades and assimilates poly(ethylene terephthalate).
|
|
71331
|
A bacterium that degrades and assimilates poly(ethylene terephthalate).
|
|
71332
|
Active Site Flexibility as a Hallmark for Efficient PET Degradation by I. sakaiensis PETase.
|
|
71333
|
Active Site Flexibility as a Hallmark for Efficient PET Degradation by I. sakaiensis PETase.
|
|
71334
|
Crystal structure and thermodynamic and kinetic stability of metagenome-derived LC-cutinase.
|
|
71335
|
Crystal structure and thermodynamic and kinetic stability of metagenome-derived LC-cutinase.
|
|
71336
|
Crystal structure and thermodynamic and kinetic stability of metagenome-derived LC-cutinase.
|
|
71337
|
Crystal structure and thermodynamic and kinetic stability of metagenome-derived LC-cutinase.
|
|
71338
|
Crystal structure and thermodynamic and kinetic stability of metagenome-derived LC-cutinase.
|
|
71339
|
Crystal structure and thermodynamic and kinetic stability of metagenome-derived LC-cutinase.
|
|
71340
|
Crystal structure and thermodynamic and kinetic stability of metagenome-derived LC-cutinase.
|
|
71341
|
Crystal structure and thermodynamic and kinetic stability of metagenome-derived LC-cutinase.
|
|
71342
|
Crystal structure and thermodynamic and kinetic stability of metagenome-derived LC-cutinase.
|
|
71343
|
Fluorinated substrates result in variable leakage of a reaction intermediate during catalysis by ...
|
|
71344
|
Fluorinated substrates result in variable leakage of a reaction intermediate during catalysis by ...
|
|
71345
|
Fluorinated substrates result in variable leakage of a reaction intermediate during catalysis by ...
|
|
71346
|
Fluorinated substrates result in variable leakage of a reaction intermediate during catalysis by ...
|
|
71347
|
Fluorinated substrates result in variable leakage of a reaction intermediate during catalysis by ...
|
|
71348
|
Fluorinated substrates result in variable leakage of a reaction intermediate during catalysis by ...
|
|
71349
|
Fluorinated substrates result in variable leakage of a reaction intermediate during catalysis by ...
|
|
71350
|
Fluorinated substrates result in variable leakage of a reaction intermediate during catalysis by ...
|
|
71351
|
Fluorinated substrates result in variable leakage of a reaction intermediate during catalysis by ...
|
|
71352
|
UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase of Escherichia coli is a zinc metalloenzyme
|
|
71353
|
UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase of Escherichia coli is a zinc metalloenzyme
|
|
71354
|
UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase of Escherichia coli is a zinc metalloenzyme
|
|
71355
|
UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase of Escherichia coli is a zinc metalloenzyme
|
|
71356
|
UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase of Escherichia coli is a zinc metalloenzyme
|
|
71357
|
UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase of Escherichia coli is a zinc metalloenzyme
|
|
71358
|
Phosphoenolpyruvate synthetase of Escherichia coli. Purification, some properties, and the role of divalent ...
|
|
71359
|
Phosphoenolpyruvate synthetase of Escherichia coli. Purification, some properties, and the role of divalent ...
|
|
71360
|
Phosphoenolpyruvate synthetase of Escherichia coli. Purification, some properties, and the role of divalent ...
|
|
71361
|
Phosphoenolpyruvate synthetase of Escherichia coli. Purification, some properties, and the role of divalent ...
|
|
71362
|
Phosphoenolpyruvate synthetase of Escherichia coli. Purification, some properties, and the role of divalent ...
|
|
71363
|
Phosphoenolpyruvate synthetase of Escherichia coli. Purification, some properties, and the role of divalent ...
|
|
71364
|
Phosphoenolpyruvate synthetase of Escherichia coli. Purification, some properties, and the role of divalent ...
|
|
71365
|
Phosphoenolpyruvate synthetase of Escherichia coli. Purification, some properties, and the role of divalent ...
|
|
71366
|
Conformational dynamics of the active site loop of S-adenosylmethionine synthetase illuminated by ...
|
|
71367
|
Conformational dynamics of the active site loop of S-adenosylmethionine synthetase illuminated by ...
|
|
71368
|
Conformational dynamics of the active site loop of S-adenosylmethionine synthetase illuminated by ...
|
|
71369
|
Conformational dynamics of the active site loop of S-adenosylmethionine synthetase illuminated by ...
|
|
71370
|
Conformational dynamics of the active site loop of S-adenosylmethionine synthetase illuminated by ...
|
|
71371
|
Mycobacterium tuberculosis pantothenate kinase: possible changes in location of ligands during enzyme action
|
|
71372
|
Mycobacterium tuberculosis pantothenate kinase: possible changes in location of ligands during enzyme action
|
|
71373
|
Mycobacterium tuberculosis pantothenate kinase: possible changes in location of ligands during enzyme action
|
|
71374
|
Mycobacterium tuberculosis pantothenate kinase: possible changes in location of ligands during enzyme action
|
|
71375
|
The role of evolutionarily conserved hydrophobic contacts in the quaternary structure stability of Escherichia ...
|
|
71376
|
The role of evolutionarily conserved hydrophobic contacts in the quaternary structure stability of Escherichia ...
|
|
71377
|
The role of evolutionarily conserved hydrophobic contacts in the quaternary structure stability of Escherichia ...
|
|
71378
|
The role of evolutionarily conserved hydrophobic contacts in the quaternary structure stability of Escherichia ...
|
|
71379
|
The role of evolutionarily conserved hydrophobic contacts in the quaternary structure stability of Escherichia ...
|
|
71380
|
The role of evolutionarily conserved hydrophobic contacts in the quaternary structure stability of Escherichia ...
|
|
71381
|
The role of evolutionarily conserved hydrophobic contacts in the quaternary structure stability of Escherichia ...
|
|
71382
|
The role of evolutionarily conserved hydrophobic contacts in the quaternary structure stability of Escherichia ...
|
|
71383
|
The role of evolutionarily conserved hydrophobic contacts in the quaternary structure stability of Escherichia ...
|
|
71384
|
The role of evolutionarily conserved hydrophobic contacts in the quaternary structure stability of Escherichia ...
|
|
71385
|
The role of evolutionarily conserved hydrophobic contacts in the quaternary structure stability of Escherichia ...
|
|
71386
|
The role of evolutionarily conserved hydrophobic contacts in the quaternary structure stability of Escherichia ...
|
|
71387
|
The role of evolutionarily conserved hydrophobic contacts in the quaternary structure stability of Escherichia ...
|
|
71388
|
The role of evolutionarily conserved hydrophobic contacts in the quaternary structure stability of Escherichia ...
|
|
71389
|
The role of evolutionarily conserved hydrophobic contacts in the quaternary structure stability of Escherichia ...
|
|
71390
|
The role of evolutionarily conserved hydrophobic contacts in the quaternary structure stability of Escherichia ...
|
|
71391
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71392
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71393
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71394
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71395
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71396
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71397
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71398
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71399
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71400
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71401
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71402
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71403
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71404
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71405
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71406
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71407
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71408
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71409
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71410
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71411
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71412
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71413
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71414
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71415
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71416
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71417
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71418
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71419
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71420
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71421
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71422
|
Site-directed fluorescence labeling reveals differences on the R-conformer of glucosamine 6-phosphate ...
|
|
71423
|
Alterations of bile acid and bumetanide uptake during culturing of rat hepatocytes.
|
|
71424
|
Alterations of bile acid and bumetanide uptake during culturing of rat hepatocytes.
|
|
71425
|
Alterations of bile acid and bumetanide uptake during culturing of rat hepatocytes.
|
|
71426
|
Different reactivity of two brain sialyltransferases towards sulfhydryl reagents. Evidence for a thiol group ...
|
|
71427
|
Different reactivity of two brain sialyltransferases towards sulfhydryl reagents. Evidence for a thiol group ...
|
|
71428
|
Different reactivity of two brain sialyltransferases towards sulfhydryl reagents. Evidence for a thiol group ...
|
|
71429
|
Different reactivity of two brain sialyltransferases towards sulfhydryl reagents. Evidence for a thiol group ...
|
|
71430
|
Different reactivity of two brain sialyltransferases towards sulfhydryl reagents. Evidence for a thiol group ...
|
|
71431
|
Different reactivity of two brain sialyltransferases towards sulfhydryl reagents. Evidence for a thiol group ...
|
|
71432
|
Different reactivity of two brain sialyltransferases towards sulfhydryl reagents. Evidence for a thiol group ...
|
|
71433
|
A novel inhibitor that suspends the induced fit mechanism of UDP-N- acetylglucosamine enolpyruvyl transferase ...
|
|
71434
|
A novel inhibitor that suspends the induced fit mechanism of UDP-N- acetylglucosamine enolpyruvyl transferase ...
|
|
71435
|
Purification and properties of L-histidine decarboxylase from mouse stomach.
|
|
71436
|
Biochemical characterization of the cutinases from Thermobifida fusca
|
|
71437
|
Biochemical characterization of the cutinases from Thermobifida fusca
|
|
71438
|
Biochemical characterization of the cutinases from Thermobifida fusca
|
|
71439
|
Biochemical characterization of the cutinases from Thermobifida fusca
|
|
71440
|
Biochemical characterization of the cutinases from Thermobifida fusca
|
|
71441
|
Biochemical characterization of the cutinases from Thermobifida fusca
|
|
71442
|
Biochemical and genetic analysis of a cutinase-type polyesterase from a thermophilic Thermobifida alba AHK119
|
|
71443
|
Biochemical and genetic analysis of a cutinase-type polyesterase from a thermophilic Thermobifida alba AHK119
|
|
71444
|
Biochemical and genetic analysis of a cutinase-type polyesterase from a thermophilic Thermobifida alba AHK119
|
|
71445
|
Biochemical and genetic analysis of a cutinase-type polyesterase from a thermophilic Thermobifida alba AHK119
|
|
71446
|
Novel antifungal drug discovery based on targeting pathways regulating the fungus-conserved Upc2 transcription ...
|
|
71447
|
Structural insights into substrate and inhibitor binding sites in human indoleamine 2,3-dioxygenase 1
|
|
71448
|
Structural insights into substrate and inhibitor binding sites in human indoleamine 2,3-dioxygenase 1
|
|
71449
|
Structural insights into substrate and inhibitor binding sites in human indoleamine 2,3-dioxygenase 1
|
|
71450
|
Structural insights into substrate and inhibitor binding sites in human indoleamine 2,3-dioxygenase 1
|
|
71451
|
Cutinase-Catalyzed Hydrolysis of Poly(ethylene terephthalate)
|
|
71452
|
Cutinase-Catalyzed Hydrolysis of Poly(ethylene terephthalate)
|
|
71453
|
Cutinase-Catalyzed Hydrolysis of Poly(ethylene terephthalate)
|
|
71454
|
Enzymatic Surface Hydrolysis of PET: Effect of Structural Diversity on Kinetic Properties of Cutinases from ...
|
|
71455
|
Enzymatic Surface Hydrolysis of PET: Effect of Structural Diversity on Kinetic Properties of Cutinases from ...
|
|
71456
|
Enzymatic Surface Hydrolysis of PET: Effect of Structural Diversity on Kinetic Properties of Cutinases from ...
|
|
71457
|
Enzymatic Surface Hydrolysis of PET: Effect of Structural Diversity on Kinetic Properties of Cutinases from ...
|
|
71458
|
Enzymatic Surface Hydrolysis of PET: Effect of Structural Diversity on Kinetic Properties of Cutinases from ...
|
|
71459
|
Enzymatic Surface Hydrolysis of PET: Effect of Structural Diversity on Kinetic Properties of Cutinases from ...
|
|
71460
|
Cloning, nucleotide sequencing and characterization of a polyurethanase gene (pueB) from Pseudomonas ...
|
|
71461
|
Cloning, nucleotide sequencing and characterization of a polyurethanase gene (pueB) from Pseudomonas ...
|
|
71462
|
Cloning, nucleotide sequencing and characterization of a polyurethanase gene (pueB) from Pseudomonas ...
|
|
71463
|
Cloning, nucleotide sequencing and characterization of a polyurethanase gene (pueB) from Pseudomonas ...
|
|
71464
|
Cloning, nucleotide sequencing and characterization of a polyurethanase gene (pueB) from Pseudomonas ...
|
|
71465
|
Lipase from Candida antarctica (CALB) and cutinase from Humicola insolens act synergistically for PET ...
|
|
71466
|
Lipase from Candida antarctica (CALB) and cutinase from Humicola insolens act synergistically for PET ...
|
|
71467
|
A New Esterase from Thermobifida halotolerans Hydrolyses Polyethylene Terephthalate (PET) and Polylactic Acid ...
|
|
71468
|
A New Esterase from Thermobifida halotolerans Hydrolyses Polyethylene Terephthalate (PET) and Polylactic Acid ...
|
|
71469
|
Cloning and expression in Escherichia coli of a polyurethane-degrading enzyme from Pseudomonas fluorescens
|
|
71470
|
Cloning and expression in Escherichia coli of a polyurethane-degrading enzyme from Pseudomonas fluorescens
|
|
71471
|
Growth of Acinetobacter gerneri P7 on polyurethane and the purification and characterization of a ...
|
|
71472
|
Growth of Acinetobacter gerneri P7 on polyurethane and the purification and characterization of a ...
|
|
71473
|
Growth of Acinetobacter gerneri P7 on polyurethane and the purification and characterization of a ...
|
|
71474
|
Growth of Acinetobacter gerneri P7 on polyurethane and the purification and characterization of a ...
|
|
71475
|
Growth of Acinetobacter gerneri P7 on polyurethane and the purification and characterization of a ...
|
|
71476
|
Growth of Acinetobacter gerneri P7 on polyurethane and the purification and characterization of a ...
|
|
71477
|
Purification and characterization of l-histidine decarboxylase from mouse mastocytoma P-815 cells.
|
|
71478
|
Spectroscopic analysis of recombinant rat histidine decarboxylase
|
|
71479
|
Growth of Pseudomonas chlororaphis on a polyester-polyurethane and the purification and characterization of a ...
|
|
71480
|
Growth of Bacillus subtilis on polyurethane and the purification and characterization of a ...
|
|
71481
|
Growth of Bacillus subtilis on polyurethane and the purification and characterization of a ...
|
|
71482
|
Growth of Bacillus subtilis on polyurethane and the purification and characterization of a ...
|
|
71483
|
Growth of Bacillus subtilis on polyurethane and the purification and characterization of a ...
|
|
71484
|
Growth of Bacillus subtilis on polyurethane and the purification and characterization of a ...
|
|
71485
|
Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch ...
|
|
71486
|
Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch ...
|
|
71487
|
Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch ...
|
|
71488
|
Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch ...
|
|
71489
|
Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch ...
|
|
71490
|
Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch ...
|
|
71491
|
Molecular insights into the pathogenicity of variants associated with the aromatic amino acid decarboxylase ...
|
|
71492
|
Molecular insights into the pathogenicity of variants associated with the aromatic amino acid decarboxylase ...
|
|
71493
|
Molecular insights into the pathogenicity of variants associated with the aromatic amino acid decarboxylase ...
|
|
71494
|
Molecular insights into the pathogenicity of variants associated with the aromatic amino acid decarboxylase ...
|
|
71495
|
Molecular insights into the pathogenicity of variants associated with the aromatic amino acid decarboxylase ...
|
|
71496
|
Molecular insights into the pathogenicity of variants associated with the aromatic amino acid decarboxylase ...
|
|
71497
|
Molecular insights into the pathogenicity of variants associated with the aromatic amino acid decarboxylase ...
|
|
71498
|
Molecular insights into the pathogenicity of variants associated with the aromatic amino acid decarboxylase ...
|
|
71499
|
Molecular insights into the pathogenicity of variants associated with the aromatic amino acid decarboxylase ...
|
|
71500
|
Molecular insights into the pathogenicity of variants associated with the aromatic amino acid decarboxylase ...
|
|
71501
|
Molecular insights into the pathogenicity of variants associated with the aromatic amino acid decarboxylase ...
|
|
71502
|
Molecular insights into the pathogenicity of variants associated with the aromatic amino acid decarboxylase ...
|
|
71503
|
Molecular insights into the pathogenicity of variants associated with the aromatic amino acid decarboxylase ...
|
|
71504
|
Molecular insights into the pathogenicity of variants associated with the aromatic amino acid decarboxylase ...
|
|
71505
|
Molecular insights into the pathogenicity of variants associated with the aromatic amino acid decarboxylase ...
|
|
71506
|
Molecular insights into the pathogenicity of variants associated with the aromatic amino acid decarboxylase ...
|
|
71507
|
Molecular insights into the pathogenicity of variants associated with the aromatic amino acid decarboxylase ...
|
|
71508
|
Molecular insights into the pathogenicity of variants associated with the aromatic amino acid decarboxylase ...
|
|
71509
|
Molecular insights into the pathogenicity of variants associated with the aromatic amino acid decarboxylase ...
|
|
71510
|
Molecular insights into the pathogenicity of variants associated with the aromatic amino acid decarboxylase ...
|
|
71511
|
Molecular insights into the pathogenicity of variants associated with the aromatic amino acid decarboxylase ...
|
|
71512
|
Molecular insights into the pathogenicity of variants associated with the aromatic amino acid decarboxylase ...
|
|
71513
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71514
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71515
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71516
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71517
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71518
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71519
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71520
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71521
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71522
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71523
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71524
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71525
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71526
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71527
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71528
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71529
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71530
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71531
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71532
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71533
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71534
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71535
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71536
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71537
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71538
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71539
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71540
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71541
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71542
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71543
|
An engineered PET depolymerase to break down and recycle plastic bottles
|
|
71544
|
Two (betaalpha)(8)-barrel enzymes of histidine and tryptophan biosynthesis have similar reaction mechanisms ...
|
|
71545
|
Two (betaalpha)(8)-barrel enzymes of histidine and tryptophan biosynthesis have similar reaction mechanisms ...
|
|
71546
|
Two (betaalpha)(8)-barrel enzymes of histidine and tryptophan biosynthesis have similar reaction mechanisms ...
|
|
71547
|
Two (betaalpha)(8)-barrel enzymes of histidine and tryptophan biosynthesis have similar reaction mechanisms ...
|
|
71548
|
Two (betaalpha)(8)-barrel enzymes of histidine and tryptophan biosynthesis have similar reaction mechanisms ...
|
|
71549
|
Two (betaalpha)(8)-barrel enzymes of histidine and tryptophan biosynthesis have similar reaction mechanisms ...
|
|
71550
|
Two (betaalpha)(8)-barrel enzymes of histidine and tryptophan biosynthesis have similar reaction mechanisms ...
|
|
71551
|
Two (betaalpha)(8)-barrel enzymes of histidine and tryptophan biosynthesis have similar reaction mechanisms ...
|
|
71552
|
Two (betaalpha)(8)-barrel enzymes of histidine and tryptophan biosynthesis have similar reaction mechanisms ...
|
|
71553
|
Two (betaalpha)(8)-barrel enzymes of histidine and tryptophan biosynthesis have similar reaction mechanisms ...
|
|
71554
|
Two (betaalpha)(8)-barrel enzymes of histidine and tryptophan biosynthesis have similar reaction mechanisms ...
|
|
71555
|
Two (betaalpha)(8)-barrel enzymes of histidine and tryptophan biosynthesis have similar reaction mechanisms ...
|
|
71556
|
Two (betaalpha)(8)-barrel enzymes of histidine and tryptophan biosynthesis have similar reaction mechanisms ...
|
|
71557
|
Kinetic and mechanistic analysis of the Escherichia coli ribD-encoded bifunctional deaminase-reductase ...
|
|
71558
|
Kinetic and mechanistic analysis of the Escherichia coli ribD-encoded bifunctional deaminase-reductase ...
|
|
71559
|
Kinetic and mechanistic analysis of the Escherichia coli ribD-encoded bifunctional deaminase-reductase ...
|
|
71560
|
Kinetic and mechanistic analysis of the Escherichia coli ribD-encoded bifunctional deaminase-reductase ...
|
|
71561
|
Kinetic and mechanistic analysis of the Escherichia coli ribD-encoded bifunctional deaminase-reductase ...
|
|
71562
|
Kinetic and mechanistic analysis of the Escherichia coli ribD-encoded bifunctional deaminase-reductase ...
|
|
71563
|
Kinetic and mechanistic analysis of the Escherichia coli ribD-encoded bifunctional deaminase-reductase ...
|
|
71564
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71565
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71566
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71567
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71568
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71569
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71570
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71571
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71572
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71573
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71574
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71575
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71576
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71577
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71578
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71579
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71580
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71581
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71582
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71583
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71584
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71585
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71586
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71587
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71588
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71589
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71590
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71591
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71592
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71593
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71594
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71595
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71596
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71597
|
Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis.
|
|
71598
|
Role of the amino acid invariants in the active site of MurG as evaluated by site-directed mutagenesis.
|
|
71599
|
Role of the amino acid invariants in the active site of MurG as evaluated by site-directed mutagenesis.
|
|
71600
|
Role of the amino acid invariants in the active site of MurG as evaluated by site-directed mutagenesis.
|
|
71601
|
Role of the amino acid invariants in the active site of MurG as evaluated by site-directed mutagenesis.
|
|
71602
|
Role of the amino acid invariants in the active site of MurG as evaluated by site-directed mutagenesis.
|
|
71603
|
Role of the amino acid invariants in the active site of MurG as evaluated by site-directed mutagenesis.
|
|
71604
|
Role of the amino acid invariants in the active site of MurG as evaluated by site-directed mutagenesis.
|
|
71605
|
Role of the amino acid invariants in the active site of MurG as evaluated by site-directed mutagenesis.
|
|
71606
|
Role of the amino acid invariants in the active site of MurG as evaluated by site-directed mutagenesis.
|
|
71607
|
Role of the amino acid invariants in the active site of MurG as evaluated by site-directed mutagenesis.
|
|
71608
|
Role of the amino acid invariants in the active site of MurG as evaluated by site-directed mutagenesis.
|
|
71609
|
Role of the amino acid invariants in the active site of MurG as evaluated by site-directed mutagenesis.
|
|
71610
|
Role of the amino acid invariants in the active site of MurG as evaluated by site-directed mutagenesis.
|
|
71611
|
Role of the amino acid invariants in the active site of MurG as evaluated by site-directed mutagenesis.
|
|
71612
|
Role of the amino acid invariants in the active site of MurG as evaluated by site-directed mutagenesis.
|
|
71613
|
Role of the amino acid invariants in the active site of MurG as evaluated by site-directed mutagenesis.
|
|
71614
|
Role of the amino acid invariants in the active site of MurG as evaluated by site-directed mutagenesis.
|
|
71615
|
Role of the amino acid invariants in the active site of MurG as evaluated by site-directed mutagenesis.
|
|
71616
|
N5-CAIR mutase: role of a CO2 binding site and substrate movement in catalysis.
|
|
71617
|
N5-CAIR mutase: role of a CO2 binding site and substrate movement in catalysis.
|
|
71618
|
N5-CAIR mutase: role of a CO2 binding site and substrate movement in catalysis.
|
|
71619
|
N5-CAIR mutase: role of a CO2 binding site and substrate movement in catalysis.
|
|
71620
|
N5-CAIR mutase: role of a CO2 binding site and substrate movement in catalysis.
|
|
71621
|
N5-CAIR mutase: role of a CO2 binding site and substrate movement in catalysis.
|
|
71622
|
N5-CAIR mutase: role of a CO2 binding site and substrate movement in catalysis.
|
|
71623
|
N5-CAIR mutase: role of a CO2 binding site and substrate movement in catalysis.
|
|
71624
|
N5-CAIR mutase: role of a CO2 binding site and substrate movement in catalysis.
|
|
71625
|
N5-CAIR mutase: role of a CO2 binding site and substrate movement in catalysis.
|
|
71626
|
N5-CAIR mutase: role of a CO2 binding site and substrate movement in catalysis.
|
|
71627
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71628
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71629
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71630
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71631
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71632
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71633
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71634
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71635
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71636
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71637
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71638
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71639
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71640
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71641
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71642
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71643
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71644
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71645
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71646
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71647
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71648
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71649
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71650
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71651
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71652
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71653
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71654
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71655
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71656
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71657
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71658
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71659
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71660
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71661
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71662
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71663
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71664
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71665
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71666
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71667
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71668
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71669
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71670
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71671
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71672
|
Thiazolopyrimidine inhibitors of 2-methylerythritol 2,4-cyclodiphosphate synthase (IspF) from Mycobacterium ...
|
|
71673
|
SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity
|
|
71674
|
SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity
|
|
71675
|
SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity
|
|
71676
|
SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity
|
|
71677
|
Increase of SARS-CoV 3CL peptidase activity due to macromolecular crowding effects in the milieu composition
|
|
71678
|
Increase of SARS-CoV 3CL peptidase activity due to macromolecular crowding effects in the milieu composition
|
|
71679
|
Increase of SARS-CoV 3CL peptidase activity due to macromolecular crowding effects in the milieu composition
|
|
71680
|
Increase of SARS-CoV 3CL peptidase activity due to macromolecular crowding effects in the milieu composition
|
|
71681
|
Increase of SARS-CoV 3CL peptidase activity due to macromolecular crowding effects in the milieu composition
|
|
71682
|
Increase of SARS-CoV 3CL peptidase activity due to macromolecular crowding effects in the milieu composition
|
|
71683
|
Increase of SARS-CoV 3CL peptidase activity due to macromolecular crowding effects in the milieu composition
|
|
71684
|
Increase of SARS-CoV 3CL peptidase activity due to macromolecular crowding effects in the milieu composition
|
|
71685
|
Increase of SARS-CoV 3CL peptidase activity due to macromolecular crowding effects in the milieu composition
|
|
71686
|
Increase of SARS-CoV 3CL peptidase activity due to macromolecular crowding effects in the milieu composition
|
|
71687
|
Increase of SARS-CoV 3CL peptidase activity due to macromolecular crowding effects in the milieu composition
|
|
71688
|
Increase of SARS-CoV 3CL peptidase activity due to macromolecular crowding effects in the milieu composition
|
|
71689
|
Increase of SARS-CoV 3CL peptidase activity due to macromolecular crowding effects in the milieu composition
|
|
71690
|
Increase of SARS-CoV 3CL peptidase activity due to macromolecular crowding effects in the milieu composition
|
|
71691
|
Increase of SARS-CoV 3CL peptidase activity due to macromolecular crowding effects in the milieu composition
|
|
71692
|
Increase of SARS-CoV 3CL peptidase activity due to macromolecular crowding effects in the milieu composition
|
|
71693
|
Increase of SARS-CoV 3CL peptidase activity due to macromolecular crowding effects in the milieu composition
|
|
71694
|
Increase of SARS-CoV 3CL peptidase activity due to macromolecular crowding effects in the milieu composition
|
|
71695
|
Alpha-keto acid dehydrogenase complexes. XX. A kinetic study of the pyruvate dehydrogenase complex from bovine ...
|
|
71696
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71697
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71698
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71699
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71700
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71701
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71702
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71703
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71704
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71705
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71706
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71707
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71708
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71709
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71710
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71711
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71712
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71713
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71714
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71715
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71716
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71717
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71718
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71719
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71720
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71721
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71722
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71723
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71724
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71725
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71726
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71727
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71728
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71729
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71730
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71731
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71732
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71733
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71734
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71735
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71736
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71737
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71738
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71739
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71740
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71741
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71742
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71743
|
Correlation between dissociation and catalysis of SARS-CoV main protease
|
|
71744
|
Conformational Flexibility of a Short Loop near the Active Site of the SARS-3CLpro is Essential to Maintain ...
|
|
71745
|
Structural basis of inhibition specificities of 3C and 3C-like proteases by zinc-coordinating and ...
|
|
71746
|
Structural basis of inhibition specificities of 3C and 3C-like proteases by zinc-coordinating and ...
|
|
71747
|
Structural basis of inhibition specificities of 3C and 3C-like proteases by zinc-coordinating and ...
|
|
71748
|
Structural basis of inhibition specificities of 3C and 3C-like proteases by zinc-coordinating and ...
|
|
71749
|
Structural basis of inhibition specificities of 3C and 3C-like proteases by zinc-coordinating and ...
|
|
71750
|
Structural basis of inhibition specificities of 3C and 3C-like proteases by zinc-coordinating and ...
|
|
71751
|
Structural basis of inhibition specificities of 3C and 3C-like proteases by zinc-coordinating and ...
|
|
71752
|
Structural basis of inhibition specificities of 3C and 3C-like proteases by zinc-coordinating and ...
|
|
71753
|
Vibrio harveyi NADPH-flavin oxidoreductase: cloning, sequencing and overexpression of the gene and ...
|
|
71754
|
Structural basis for catalytic and inhibitory mechanisms of human prostaglandin reductase PTGR2
|
|
71755
|
Structural basis for catalytic and inhibitory mechanisms of human prostaglandin reductase PTGR2
|
|
71756
|
Structural basis for catalytic and inhibitory mechanisms of human prostaglandin reductase PTGR2
|
|
71757
|
Structural basis for catalytic and inhibitory mechanisms of human prostaglandin reductase PTGR2
|
|
71758
|
Structural basis for catalytic and inhibitory mechanisms of human prostaglandin reductase PTGR2
|
|
71759
|
Structural basis for catalytic and inhibitory mechanisms of human prostaglandin reductase PTGR2
|
|
71760
|
Structural basis for catalytic and inhibitory mechanisms of human prostaglandin reductase PTGR2
|
|
71761
|
A pathogenic fungi diphenyl ether phytotoxin targets plant enoyl (acyl carrier protein) reductase
|
|
71762
|
A pathogenic fungi diphenyl ether phytotoxin targets plant enoyl (acyl carrier protein) reductase
|
|
71763
|
A pathogenic fungi diphenyl ether phytotoxin targets plant enoyl (acyl carrier protein) reductase
|
|
71764
|
Studies of human 2,4-dienoyl CoA reductase shed new light on peroxisomal ?-oxidation of unsaturated fatty ...
|
|
71765
|
Studies of human 2,4-dienoyl CoA reductase shed new light on peroxisomal ?-oxidation of unsaturated fatty ...
|
|
71766
|
Structural enzymological studies of 2-enoyl thioester reductase of the human mitochondrial FAS II pathway: new ...
|
|
71767
|
Structural enzymological studies of 2-enoyl thioester reductase of the human mitochondrial FAS II pathway: new ...
|
|
71768
|
Structural enzymological studies of 2-enoyl thioester reductase of the human mitochondrial FAS II pathway: new ...
|
|
71769
|
Structural enzymological studies of 2-enoyl thioester reductase of the human mitochondrial FAS II pathway: new ...
|
|
71770
|
Structural enzymological studies of 2-enoyl thioester reductase of the human mitochondrial FAS II pathway: new ...
|
|
71771
|
Structural enzymological studies of 2-enoyl thioester reductase of the human mitochondrial FAS II pathway: new ...
|
|
71772
|
Structural enzymological studies of 2-enoyl thioester reductase of the human mitochondrial FAS II pathway: new ...
|
|
71773
|
Structural enzymological studies of 2-enoyl thioester reductase of the human mitochondrial FAS II pathway: new ...
|
|
71774
|
Structural enzymological studies of 2-enoyl thioester reductase of the human mitochondrial FAS II pathway: new ...
|
|
71775
|
Structural enzymological studies of 2-enoyl thioester reductase of the human mitochondrial FAS II pathway: new ...
|
|
71776
|
Structural enzymological studies of 2-enoyl thioester reductase of the human mitochondrial FAS II pathway: new ...
|
|
71777
|
Structural enzymological studies of 2-enoyl thioester reductase of the human mitochondrial FAS II pathway: new ...
|
|
71778
|
Structural enzymological studies of 2-enoyl thioester reductase of the human mitochondrial FAS II pathway: new ...
|
|
71779
|
Structural enzymological studies of 2-enoyl thioester reductase of the human mitochondrial FAS II pathway: new ...
|
|
71780
|
Structural enzymological studies of 2-enoyl thioester reductase of the human mitochondrial FAS II pathway: new ...
|
|
71781
|
Structural enzymological studies of 2-enoyl thioester reductase of the human mitochondrial FAS II pathway: new ...
|
|
71782
|
Structural enzymological studies of 2-enoyl thioester reductase of the human mitochondrial FAS II pathway: new ...
|
|
71783
|
Structural enzymological studies of 2-enoyl thioester reductase of the human mitochondrial FAS II pathway: new ...
|
|
71784
|
Structural enzymological studies of 2-enoyl thioester reductase of the human mitochondrial FAS II pathway: new ...
|
|
71785
|
Structural enzymological studies of 2-enoyl thioester reductase of the human mitochondrial FAS II pathway: new ...
|
|
71786
|
Structural enzymological studies of 2-enoyl thioester reductase of the human mitochondrial FAS II pathway: new ...
|
|
71787
|
Purification, characterization, and molecular cloning of S-adenosyl-L-methionine: uroporphyrinogen III ...
|
|
71788
|
The Akt C-terminal modulator protein is an acyl-CoA thioesterase of the Hotdog-Fold family
|
|
71789
|
The Akt C-terminal modulator protein is an acyl-CoA thioesterase of the Hotdog-Fold family
|
|
71790
|
The Akt C-terminal modulator protein is an acyl-CoA thioesterase of the Hotdog-Fold family
|
|
71791
|
The Akt C-terminal modulator protein is an acyl-CoA thioesterase of the Hotdog-Fold family
|
|
71792
|
The Akt C-terminal modulator protein is an acyl-CoA thioesterase of the Hotdog-Fold family
|
|
71793
|
The Akt C-terminal modulator protein is an acyl-CoA thioesterase of the Hotdog-Fold family
|
|
71794
|
The Akt C-terminal modulator protein is an acyl-CoA thioesterase of the Hotdog-Fold family
|
|
71795
|
The Akt C-terminal modulator protein is an acyl-CoA thioesterase of the Hotdog-Fold family
|
|
71796
|
The Akt C-terminal modulator protein is an acyl-CoA thioesterase of the Hotdog-Fold family
|
|
71797
|
The Akt C-terminal modulator protein is an acyl-CoA thioesterase of the Hotdog-Fold family
|
|
71798
|
The Akt C-terminal modulator protein is an acyl-CoA thioesterase of the Hotdog-Fold family
|
|
71799
|
The Akt C-terminal modulator protein is an acyl-CoA thioesterase of the Hotdog-Fold family
|
|
71800
|
The Akt C-terminal modulator protein is an acyl-CoA thioesterase of the Hotdog-Fold family
|
|
71801
|
The Akt C-terminal modulator protein is an acyl-CoA thioesterase of the Hotdog-Fold family
|
|
71802
|
The Akt C-terminal modulator protein is an acyl-CoA thioesterase of the Hotdog-Fold family
|
|
71803
|
The Akt C-terminal modulator protein is an acyl-CoA thioesterase of the Hotdog-Fold family
|
|
71804
|
The Akt C-terminal modulator protein is an acyl-CoA thioesterase of the Hotdog-Fold family
|
|
71805
|
The Akt C-terminal modulator protein is an acyl-CoA thioesterase of the Hotdog-Fold family
|
|
71806
|
The Akt C-terminal modulator protein is an acyl-CoA thioesterase of the Hotdog-Fold family
|
|
71807
|
The Akt C-terminal modulator protein is an acyl-CoA thioesterase of the Hotdog-Fold family
|
|
71808
|
The Akt C-terminal modulator protein is an acyl-CoA thioesterase of the Hotdog-Fold family
|
|
71809
|
The Akt C-terminal modulator protein is an acyl-CoA thioesterase of the Hotdog-Fold family
|
|
71810
|
The Akt C-terminal modulator protein is an acyl-CoA thioesterase of the Hotdog-Fold family
|
|
71811
|
Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double ...
|
|
71812
|
Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double ...
|
|
71813
|
Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double ...
|
|
71814
|
Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, ...
|
|
71815
|
Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, ...
|
|
71816
|
Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, ...
|
|
71817
|
Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, ...
|
|
71818
|
Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, ...
|
|
71819
|
Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, ...
|
|
71820
|
Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, ...
|
|
71821
|
Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, ...
|
|
71822
|
Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, ...
|
|
71823
|
Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, ...
|
|
71824
|
Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, ...
|
|
71825
|
Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, ...
|
|
71826
|
Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, ...
|
|
71827
|
Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, ...
|
|
71828
|
Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, ...
|
|
71829
|
Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, ...
|
|
71830
|
Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, ...
|
|
71831
|
Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, ...
|
|
71832
|
Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, ...
|
|
71833
|
Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, ...
|
|
71834
|
Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, ...
|
|
71835
|
Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, ...
|
|
71836
|
Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, ...
|
|
71837
|
Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, ...
|
|
71838
|
Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, ...
|
|
71839
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71840
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71841
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71842
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71843
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71844
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71845
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71846
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71847
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71848
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71849
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71850
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71851
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71852
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71853
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71854
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71855
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71856
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71857
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71858
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71859
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71860
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71861
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71862
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71863
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71864
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71865
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71866
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71867
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71868
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71869
|
Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: ...
|
|
71870
|
A highly charged region in the middle domain of plant endoplasmic reticulum (ER)-localized heat-shock protein ...
|
|
71871
|
A highly charged region in the middle domain of plant endoplasmic reticulum (ER)-localized heat-shock protein ...
|
|
71872
|
A highly charged region in the middle domain of plant endoplasmic reticulum (ER)-localized heat-shock protein ...
|
|
71873
|
A highly charged region in the middle domain of plant endoplasmic reticulum (ER)-localized heat-shock protein ...
|
|
71874
|
The gamma-aminobutyrate permease GabP serves as the third proline transporter of Bacillus subtilis.
|
|
71875
|
The gamma-aminobutyrate permease GabP serves as the third proline transporter of Bacillus subtilis.
|
|
71876
|
The gamma-aminobutyrate permease GabP serves as the third proline transporter of Bacillus subtilis.
|
|
71877
|
Identification and Functional Assessment of a New CYP2C9 Allelic Variant CYP2C9*59.
|
|
71878
|
Identification and Functional Assessment of a New CYP2C9 Allelic Variant CYP2C9*59.
|
|
71879
|
Identification and Functional Assessment of a New CYP2C9 Allelic Variant CYP2C9*59.
|
|
71880
|
Identification and Functional Assessment of a New CYP2C9 Allelic Variant CYP2C9*59.
|
|
71881
|
Identification and Functional Assessment of a New CYP2C9 Allelic Variant CYP2C9*59.
|
|
71882
|
Identification and Functional Assessment of a New CYP2C9 Allelic Variant CYP2C9*59.
|
|
71883
|
Identification and Functional Assessment of a New CYP2C9 Allelic Variant CYP2C9*59.
|
|
71884
|
Identification and Functional Assessment of a New CYP2C9 Allelic Variant CYP2C9*59.
|
|
71885
|
Identification and Functional Assessment of a New CYP2C9 Allelic Variant CYP2C9*59.
|
|
71886
|
Association of both enoyl coenzyme A hydratase and 3-hydroxyacyl coenzyme A epimerase with an active site in ...
|
|
71887
|
Association of both enoyl coenzyme A hydratase and 3-hydroxyacyl coenzyme A epimerase with an active site in ...
|
|
71888
|
Association of both enoyl coenzyme A hydratase and 3-hydroxyacyl coenzyme A epimerase with an active site in ...
|
|
71889
|
Association of both enoyl coenzyme A hydratase and 3-hydroxyacyl coenzyme A epimerase with an active site in ...
|
|
71890
|
Association of both enoyl coenzyme A hydratase and 3-hydroxyacyl coenzyme A epimerase with an active site in ...
|
|
71891
|
Association of both enoyl coenzyme A hydratase and 3-hydroxyacyl coenzyme A epimerase with an active site in ...
|
|
71892
|
Association of both enoyl coenzyme A hydratase and 3-hydroxyacyl coenzyme A epimerase with an active site in ...
|
|
71893
|
Association of both enoyl coenzyme A hydratase and 3-hydroxyacyl coenzyme A epimerase with an active site in ...
|
|
71894
|
Stable benzotriazole esters as mechanism-based inactivators of the severe acute respiratory syndrome 3CL ...
|
|
71895
|
Stable benzotriazole esters as mechanism-based inactivators of the severe acute respiratory syndrome 3CL ...
|
|
71896
|
Stable benzotriazole esters as mechanism-based inactivators of the severe acute respiratory syndrome 3CL ...
|
|
71897
|
Stable benzotriazole esters as mechanism-based inactivators of the severe acute respiratory syndrome 3CL ...
|
|
71898
|
Stable benzotriazole esters as mechanism-based inactivators of the severe acute respiratory syndrome 3CL ...
|
|
71899
|
Stable benzotriazole esters as mechanism-based inactivators of the severe acute respiratory syndrome 3CL ...
|
|
71900
|
Stable benzotriazole esters as mechanism-based inactivators of the severe acute respiratory syndrome 3CL ...
|
|
71901
|
Stable benzotriazole esters as mechanism-based inactivators of the severe acute respiratory syndrome 3CL ...
|
|
71902
|
Stable benzotriazole esters as mechanism-based inactivators of the severe acute respiratory syndrome 3CL ...
|
|
71903
|
Stable benzotriazole esters as mechanism-based inactivators of the severe acute respiratory syndrome 3CL ...
|
|
71904
|
Stable benzotriazole esters as mechanism-based inactivators of the severe acute respiratory syndrome 3CL ...
|
|
71905
|
Engineering a novel endopeptidase based on SARS 3CL(pro).
|
|
71906
|
Engineering a novel endopeptidase based on SARS 3CL(pro).
|
|
71907
|
Phosphatidic acid binds and stimulates Arabidopsis sphingosine kinases.
|
|
71908
|
Phosphatidic acid binds and stimulates Arabidopsis sphingosine kinases.
|
|
71909
|
Phosphatidic acid binds and stimulates Arabidopsis sphingosine kinases.
|
|
71910
|
Phosphatidic acid binds and stimulates Arabidopsis sphingosine kinases.
|
|
71911
|
Phosphatidic acid binds and stimulates Arabidopsis sphingosine kinases.
|
|
71912
|
Phosphatidic acid binds and stimulates Arabidopsis sphingosine kinases.
|
|
71913
|
Phosphatidic acid binds and stimulates Arabidopsis sphingosine kinases.
|
|
71914
|
Phosphatidic acid binds and stimulates Arabidopsis sphingosine kinases.
|
|
71915
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71916
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71917
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71918
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71919
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71920
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71921
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71922
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71923
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71924
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71925
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71926
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71927
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71928
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71929
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71930
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71931
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71932
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71933
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71934
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71935
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71936
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71937
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71938
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71939
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71940
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71941
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71942
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71943
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71944
|
Molecular basis of perhydrolase activity in serine hydrolases
|
|
71945
|
Enzymatic activity of the SARS coronavirus main proteinase dimer
|
|
71946
|
pH-dependent conformational flexibility of the SARS-CoV main proteinase (M(pro)) dimer: molecular dynamics ...
|
|
71947
|
The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen ...
|
|
71948
|
The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen ...
|
|
71949
|
Insight into the chemistry of flavin reduction and oxidation in Escherichia coli dihydroorotate dehydrogenase ...
|
|
71950
|
Insight into the chemistry of flavin reduction and oxidation in Escherichia coli dihydroorotate dehydrogenase ...
|
|
71951
|
Insight into the chemistry of flavin reduction and oxidation in Escherichia coli dihydroorotate dehydrogenase ...
|
|
71952
|
Insight into the chemistry of flavin reduction and oxidation in Escherichia coli dihydroorotate dehydrogenase ...
|
|
71953
|
Insight into the chemistry of flavin reduction and oxidation in Escherichia coli dihydroorotate dehydrogenase ...
|
|
71954
|
Insight into the chemistry of flavin reduction and oxidation in Escherichia coli dihydroorotate dehydrogenase ...
|
|
71955
|
Insight into the chemistry of flavin reduction and oxidation in Escherichia coli dihydroorotate dehydrogenase ...
|
|
71956
|
Insight into the chemistry of flavin reduction and oxidation in Escherichia coli dihydroorotate dehydrogenase ...
|
|
71957
|
Insight into the chemistry of flavin reduction and oxidation in Escherichia coli dihydroorotate dehydrogenase ...
|
|
71958
|
Insight into the chemistry of flavin reduction and oxidation in Escherichia coli dihydroorotate dehydrogenase ...
|
|
71959
|
Insight into the chemistry of flavin reduction and oxidation in Escherichia coli dihydroorotate dehydrogenase ...
|
|
71960
|
Functional characterization of protein variants encoded by nonsynonymous single nucleotide polymorphisms in ...
|
|
71961
|
Functional characterization of protein variants encoded by nonsynonymous single nucleotide polymorphisms in ...
|
|
71962
|
Functional characterization of protein variants encoded by nonsynonymous single nucleotide polymorphisms in ...
|
|
71963
|
Functional characterization of protein variants encoded by nonsynonymous single nucleotide polymorphisms in ...
|
|
71964
|
Functional characterization of protein variants encoded by nonsynonymous single nucleotide polymorphisms in ...
|
|
71965
|
Functional characterization of protein variants encoded by nonsynonymous single nucleotide polymorphisms in ...
|
|
71966
|
Functional characterization of protein variants encoded by nonsynonymous single nucleotide polymorphisms in ...
|
|
71967
|
Functional characterization of protein variants encoded by nonsynonymous single nucleotide polymorphisms in ...
|
|
71968
|
Functional characterization of protein variants encoded by nonsynonymous single nucleotide polymorphisms in ...
|
|
71969
|
Functional characterization of protein variants encoded by nonsynonymous single nucleotide polymorphisms in ...
|
|
71970
|
Functional characterization of protein variants encoded by nonsynonymous single nucleotide polymorphisms in ...
|
|
71971
|
Functional characterization of protein variants encoded by nonsynonymous single nucleotide polymorphisms in ...
|
|
71972
|
Functional characterization of protein variants encoded by nonsynonymous single nucleotide polymorphisms in ...
|
|
71973
|
Functional characterization of protein variants encoded by nonsynonymous single nucleotide polymorphisms in ...
|
|
71974
|
Functional characterization of protein variants encoded by nonsynonymous single nucleotide polymorphisms in ...
|
|
71975
|
Functional characterization of protein variants encoded by nonsynonymous single nucleotide polymorphisms in ...
|
|
71976
|
Functional characterization of protein variants encoded by nonsynonymous single nucleotide polymorphisms in ...
|
|
71977
|
Biochemical characterization of the phosphatase domain of the tumor suppressor PH domain leucine-rich repeat ...
|
|
71978
|
Biochemical characterization of the phosphatase domain of the tumor suppressor PH domain leucine-rich repeat ...
|
|
71979
|
Biochemical characterization of the phosphatase domain of the tumor suppressor PH domain leucine-rich repeat ...
|
|
71980
|
Biochemical characterization of the phosphatase domain of the tumor suppressor PH domain leucine-rich repeat ...
|
|
71981
|
Biochemical characterization of the phosphatase domain of the tumor suppressor PH domain leucine-rich repeat ...
|
|
71982
|
Biochemical characterization of the phosphatase domain of the tumor suppressor PH domain leucine-rich repeat ...
|
|
71983
|
Biochemical characterization of the phosphatase domain of the tumor suppressor PH domain leucine-rich repeat ...
|
|
71984
|
Biochemical characterization of the phosphatase domain of the tumor suppressor PH domain leucine-rich repeat ...
|
|
71985
|
Biochemical characterization of the phosphatase domain of the tumor suppressor PH domain leucine-rich repeat ...
|
|
71986
|
Biochemical characterization of the phosphatase domain of the tumor suppressor PH domain leucine-rich repeat ...
|
|
71987
|
Biochemical characterization of the phosphatase domain of the tumor suppressor PH domain leucine-rich repeat ...
|
|
71988
|
Biochemical characterization of the phosphatase domain of the tumor suppressor PH domain leucine-rich repeat ...
|
|
71989
|
The RNA acetyltransferase driven by ATP hydrolysis synthesizes N4-acetylcytidine of tRNA anticodon.
|
|
71990
|
The RNA acetyltransferase driven by ATP hydrolysis synthesizes N4-acetylcytidine of tRNA anticodon.
|
|
71991
|
The RNA acetyltransferase driven by ATP hydrolysis synthesizes N4-acetylcytidine of tRNA anticodon.
|
|
71992
|
The RNA acetyltransferase driven by ATP hydrolysis synthesizes N4-acetylcytidine of tRNA anticodon.
|
|
71993
|
The RNA acetyltransferase driven by ATP hydrolysis synthesizes N4-acetylcytidine of tRNA anticodon.
|
|
71994
|
The RNA acetyltransferase driven by ATP hydrolysis synthesizes N4-acetylcytidine of tRNA anticodon.
|
|
71995
|
The RNA acetyltransferase driven by ATP hydrolysis synthesizes N4-acetylcytidine of tRNA anticodon.
|
|
71996
|
The RNA acetyltransferase driven by ATP hydrolysis synthesizes N4-acetylcytidine of tRNA anticodon.
|
|
71997
|
Characterization of human fructose-1,6-bisphosphatase in control and deficient tissues.
|
|
71998
|
Characterization of human fructose-1,6-bisphosphatase in control and deficient tissues.
|
|
71999
|
Characterization of human fructose-1,6-bisphosphatase in control and deficient tissues.
|
|
72000
|
Characterization of human fructose-1,6-bisphosphatase in control and deficient tissues.
|





 If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.
If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.