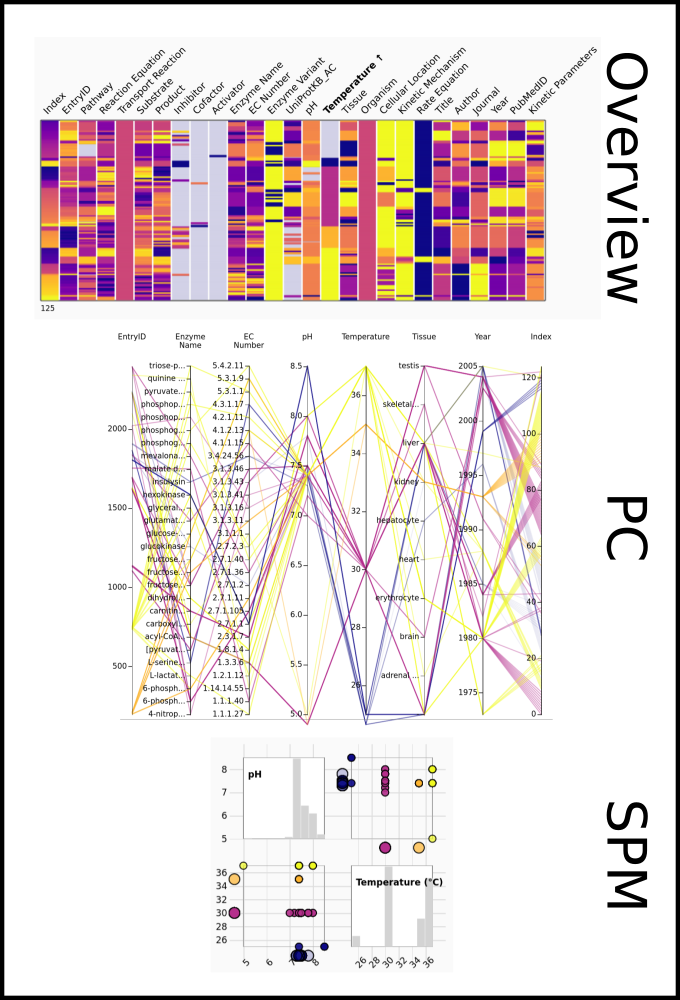
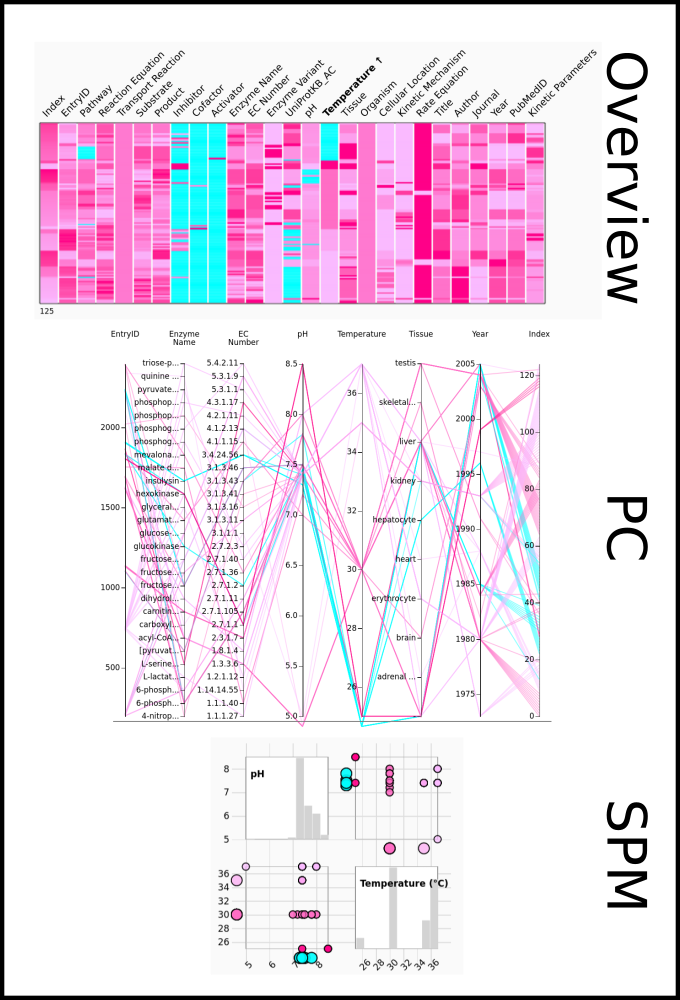
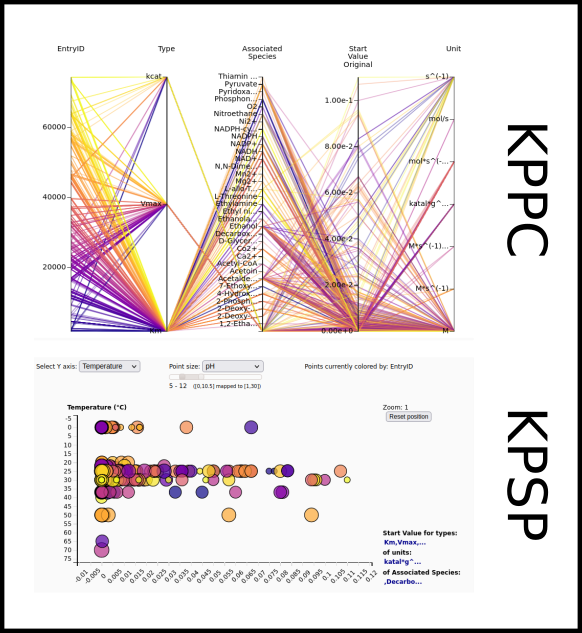
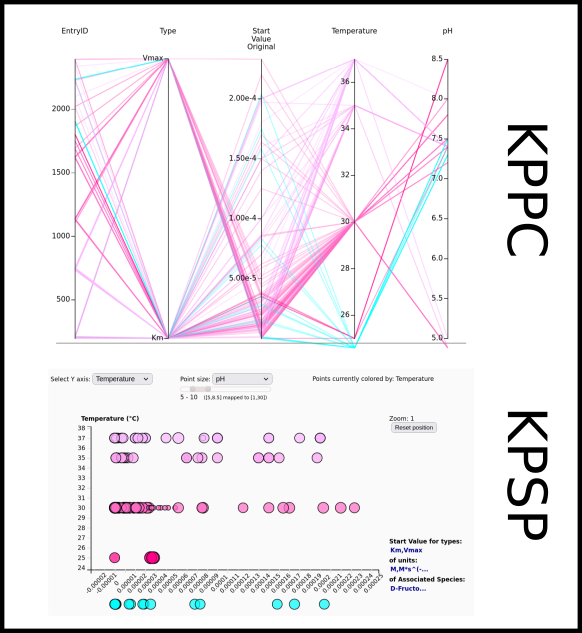
To improve SABIO-RK data interoperability, semantic markup was added to web pages as described and defined by
This structured information makes it easier to discover, collate and analyse our data.
|
7001
|
Detection of kinetically abnormal argininosuccinate synthase in neonatal citrullinemia by conversion of ...
|
|
7002
|
Detection of kinetically abnormal argininosuccinate synthase in neonatal citrullinemia by conversion of ...
|
|
7003
|
Detection of kinetically abnormal argininosuccinate synthase in neonatal citrullinemia by conversion of ...
|
|
7004
|
Detection of kinetically abnormal argininosuccinate synthase in neonatal citrullinemia by conversion of ...
|
|
7005
|
Detection of kinetically abnormal argininosuccinate synthase in neonatal citrullinemia by conversion of ...
|
|
7006
|
Detection of kinetically abnormal argininosuccinate synthase in neonatal citrullinemia by conversion of ...
|
|
7007
|
Detection of kinetically abnormal argininosuccinate synthase in neonatal citrullinemia by conversion of ...
|
|
7008
|
Detection of kinetically abnormal argininosuccinate synthase in neonatal citrullinemia by conversion of ...
|
|
7009
|
Detection of kinetically abnormal argininosuccinate synthase in neonatal citrullinemia by conversion of ...
|
|
7010
|
Detection of kinetically abnormal argininosuccinate synthase in neonatal citrullinemia by conversion of ...
|
|
7011
|
Detection of kinetically abnormal argininosuccinate synthase in neonatal citrullinemia by conversion of ...
|
|
7012
|
Detection of kinetically abnormal argininosuccinate synthase in neonatal citrullinemia by conversion of ...
|
|
7013
|
Detection of kinetically abnormal argininosuccinate synthase in neonatal citrullinemia by conversion of ...
|
|
7014
|
Detection of kinetically abnormal argininosuccinate synthase in neonatal citrullinemia by conversion of ...
|
|
7015
|
Detection of kinetically abnormal argininosuccinate synthase in neonatal citrullinemia by conversion of ...
|
|
7016
|
Detection of kinetically abnormal argininosuccinate synthase in neonatal citrullinemia by conversion of ...
|
|
7017
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7018
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7019
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7020
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7021
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7022
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7023
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7024
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7025
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7026
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7027
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7028
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7029
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7030
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7031
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7032
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7033
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7034
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7035
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7036
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7037
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7038
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7039
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7040
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7041
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7042
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7043
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7044
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7045
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7046
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7047
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7048
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7049
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7050
|
Effects of Self-Association of Ornithine Aminotransferase on its Physicochemical Characteristics
|
|
7051
|
Kinetic and Crystallographic Studies on the Active Site Arg289Lys Mutant of Flavocytochrome b2 (Yeast ...
|
|
7052
|
Kinetic and Crystallographic Studies on the Active Site Arg289Lys Mutant of Flavocytochrome b2 (Yeast ...
|
|
7053
|
Kinetic and Crystallographic Studies on the Active Site Arg289Lys Mutant of Flavocytochrome b2 (Yeast ...
|
|
7054
|
Kinetic and Crystallographic Studies on the Active Site Arg289Lys Mutant of Flavocytochrome b2 (Yeast ...
|
|
7055
|
Kinetic and Crystallographic Studies on the Active Site Arg289Lys Mutant of Flavocytochrome b2 (Yeast ...
|
|
7056
|
Kinetic and Crystallographic Studies on the Active Site Arg289Lys Mutant of Flavocytochrome b2 (Yeast ...
|
|
7057
|
Kinetic and Crystallographic Studies on the Active Site Arg289Lys Mutant of Flavocytochrome b2 (Yeast ...
|
|
7058
|
Kinetic and Crystallographic Studies on the Active Site Arg289Lys Mutant of Flavocytochrome b2 (Yeast ...
|
|
7059
|
Kinetic and Crystallographic Studies on the Active Site Arg289Lys Mutant of Flavocytochrome b2 (Yeast ...
|
|
7060
|
Effect of 6-phosphogluconate on phosphoglucose isomerase in rat brain in vitro and in vivo
|
|
7061
|
Effect of 6-phosphogluconate on phosphoglucose isomerase in rat brain in vitro and in vivo
|
|
7062
|
Effect of 6-phosphogluconate on phosphoglucose isomerase in rat brain in vitro and in vivo
|
|
7063
|
Effect of 6-phosphogluconate on phosphoglucose isomerase in rat brain in vitro and in vivo
|
|
7064
|
The putative catalytic bases have, at most, an accessory role in the mechanism of arginine kinase
|
|
7065
|
The putative catalytic bases have, at most, an accessory role in the mechanism of arginine kinase
|
|
7066
|
The putative catalytic bases have, at most, an accessory role in the mechanism of arginine kinase
|
|
7067
|
The putative catalytic bases have, at most, an accessory role in the mechanism of arginine kinase
|
|
7068
|
The putative catalytic bases have, at most, an accessory role in the mechanism of arginine kinase
|
|
7069
|
The putative catalytic bases have, at most, an accessory role in the mechanism of arginine kinase
|
|
7070
|
The putative catalytic bases have, at most, an accessory role in the mechanism of arginine kinase
|
|
7071
|
The putative catalytic bases have, at most, an accessory role in the mechanism of arginine kinase
|
|
7072
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7073
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7074
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7075
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7076
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7077
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7078
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7079
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7080
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7081
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7082
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7083
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7084
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7085
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7086
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7087
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7088
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7089
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7090
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7091
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7092
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7093
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7094
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7095
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7096
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7097
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7098
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7099
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7100
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7101
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7102
|
Identification of the catalytic residues in family 52 glycoside hydrolase, a beta-xylosidase from Geobacillus ...
|
|
7103
|
Enolase from spores and cells of Bacillus megaterium: two-step purification of the enzyme and some of its ...
|
|
7104
|
Substrate-induced inactivation of argininosuccinate lyase by monofluorofumarate and difluorofumarate
|
|
7105
|
Substrate-induced inactivation of argininosuccinate lyase by monofluorofumarate and difluorofumarate
|
|
7106
|
Substrate-induced inactivation of argininosuccinate lyase by monofluorofumarate and difluorofumarate
|
|
7107
|
13C and 15N nuclear magnetic resonance evidence of the ionization state of substrates bound to bovine ...
|
|
7108
|
13C and 15N nuclear magnetic resonance evidence of the ionization state of substrates bound to bovine ...
|
|
7109
|
13C and 15N nuclear magnetic resonance evidence of the ionization state of substrates bound to bovine ...
|
|
7110
|
13C and 15N nuclear magnetic resonance evidence of the ionization state of substrates bound to bovine ...
|
|
7111
|
Characterization and enzyme activity of argininosuccinate lyase/delta-crystallin of the embryonic duck lens
|
|
7112
|
Characterization and enzyme activity of argininosuccinate lyase/delta-crystallin of the embryonic duck lens
|
|
7113
|
Characterization and enzyme activity of argininosuccinate lyase/delta-crystallin of the embryonic duck lens
|
|
7114
|
Characterization and enzyme activity of argininosuccinate lyase/delta-crystallin of the embryonic duck lens
|
|
7115
|
The kinetic effects of in vitro phosphorylation of rabbit muscle enolase by protein kinase C. A possible new ...
|
|
7116
|
The kinetic effects of in vitro phosphorylation of rabbit muscle enolase by protein kinase C. A possible new ...
|
|
7117
|
The kinetic effects of in vitro phosphorylation of rabbit muscle enolase by protein kinase C. A possible new ...
|
|
7118
|
The kinetic effects of in vitro phosphorylation of rabbit muscle enolase by protein kinase C. A possible new ...
|
|
7119
|
Identification and characterization of differentially active pools of type IIalpha phosphatidylinositol ...
|
|
7120
|
Identification and characterization of differentially active pools of type IIalpha phosphatidylinositol ...
|
|
7121
|
Modulation of the Activity of Rat Liver Acetylglutamate Synthase by pH and Arginine Concentration
|
|
7122
|
Modulation of the Activity of Rat Liver Acetylglutamate Synthase by pH and Arginine Concentration
|
|
7123
|
Modulation of the Activity of Rat Liver Acetylglutamate Synthase by pH and Arginine Concentration
|
|
7124
|
pH-Sensitive Control of Arginase by Mn(II) Ions at Submicromolar Concentrations
|
|
7125
|
pH-Sensitive Control of Arginase by Mn(II) Ions at Submicromolar Concentrations
|
|
7126
|
pH-Sensitive Control of Arginase by Mn(II) Ions at Submicromolar Concentrations
|
|
7127
|
Subcloning of the enterobactin biosynthetic gene entB: expression, purification, characterization, and ...
|
|
7128
|
Subcloning of the enterobactin biosynthetic gene entB: expression, purification, characterization, and ...
|
|
7129
|
Subcloning of the enterobactin biosynthetic gene entB: expression, purification, characterization, and ...
|
|
7130
|
Subcloning of the enterobactin biosynthetic gene entB: expression, purification, characterization, and ...
|
|
7131
|
Subcloning of the enterobactin biosynthetic gene entB: expression, purification, characterization, and ...
|
|
7132
|
Subcloning of the enterobactin biosynthetic gene entB: expression, purification, characterization, and ...
|
|
7133
|
Subcloning of the enterobactin biosynthetic gene entB: expression, purification, characterization, and ...
|
|
7134
|
Further kinetic characterization of the non-allosteric phosphofructokinase from Escherichia coli K-12
|
|
7135
|
Further kinetic characterization of the non-allosteric phosphofructokinase from Escherichia coli K-12
|
|
7136
|
Further kinetic characterization of the non-allosteric phosphofructokinase from Escherichia coli K-12
|
|
7137
|
Further kinetic characterization of the non-allosteric phosphofructokinase from Escherichia coli K-12
|
|
7138
|
Further kinetic characterization of the non-allosteric phosphofructokinase from Escherichia coli K-12
|
|
7139
|
Further kinetic characterization of the non-allosteric phosphofructokinase from Escherichia coli K-12
|
|
7140
|
Further kinetic characterization of the non-allosteric phosphofructokinase from Escherichia coli K-12
|
|
7141
|
Further kinetic characterization of the non-allosteric phosphofructokinase from Escherichia coli K-12
|
|
7142
|
Further kinetic characterization of the non-allosteric phosphofructokinase from Escherichia coli K-12
|
|
7143
|
Further kinetic characterization of the non-allosteric phosphofructokinase from Escherichia coli K-12
|
|
7144
|
Further kinetic characterization of the non-allosteric phosphofructokinase from Escherichia coli K-12
|
|
7145
|
Further kinetic characterization of the non-allosteric phosphofructokinase from Escherichia coli K-12
|
|
7146
|
Further kinetic characterization of the non-allosteric phosphofructokinase from Escherichia coli K-12
|
|
7147
|
Further kinetic characterization of the non-allosteric phosphofructokinase from Escherichia coli K-12
|
|
7148
|
Further kinetic characterization of the non-allosteric phosphofructokinase from Escherichia coli K-12
|
|
7149
|
Further kinetic characterization of the non-allosteric phosphofructokinase from Escherichia coli K-12
|
|
7150
|
Further kinetic characterization of the non-allosteric phosphofructokinase from Escherichia coli K-12
|
|
7151
|
Further kinetic characterization of the non-allosteric phosphofructokinase from Escherichia coli K-12
|
|
7152
|
Further kinetic characterization of the non-allosteric phosphofructokinase from Escherichia coli K-12
|
|
7153
|
Further kinetic characterization of the non-allosteric phosphofructokinase from Escherichia coli K-12
|
|
7154
|
Further kinetic characterization of the non-allosteric phosphofructokinase from Escherichia coli K-12
|
|
7155
|
Further kinetic characterization of the non-allosteric phosphofructokinase from Escherichia coli K-12
|
|
7156
|
Further kinetic characterization of the non-allosteric phosphofructokinase from Escherichia coli K-12
|
|
7157
|
Further kinetic characterization of the non-allosteric phosphofructokinase from Escherichia coli K-12
|
|
7158
|
Further kinetic characterization of the non-allosteric phosphofructokinase from Escherichia coli K-12
|
|
7159
|
Further kinetic characterization of the non-allosteric phosphofructokinase from Escherichia coli K-12
|
|
7160
|
Further kinetic characterization of the non-allosteric phosphofructokinase from Escherichia coli K-12
|
|
7161
|
Glutamate 325 is a General Acid-Base Catalyst in the Reaction Catalyzed by Fructose-2,6-bisphosphatase
|
|
7162
|
Glutamate 325 is a General Acid-Base Catalyst in the Reaction Catalyzed by Fructose-2,6-bisphosphatase
|
|
7163
|
Glutamate 325 is a General Acid-Base Catalyst in the Reaction Catalyzed by Fructose-2,6-bisphosphatase
|
|
7164
|
Glutamate 325 is a General Acid-Base Catalyst in the Reaction Catalyzed by Fructose-2,6-bisphosphatase
|
|
7165
|
Glutamate 325 is a General Acid-Base Catalyst in the Reaction Catalyzed by Fructose-2,6-bisphosphatase
|
|
7166
|
Glutamate 325 is a General Acid-Base Catalyst in the Reaction Catalyzed by Fructose-2,6-bisphosphatase
|
|
7167
|
Glutamate 325 is a General Acid-Base Catalyst in the Reaction Catalyzed by Fructose-2,6-bisphosphatase
|
|
7168
|
Glutamate 325 is a General Acid-Base Catalyst in the Reaction Catalyzed by Fructose-2,6-bisphosphatase
|
|
7169
|
Glutamate 325 is a General Acid-Base Catalyst in the Reaction Catalyzed by Fructose-2,6-bisphosphatase
|
|
7170
|
Glutamate 325 is a General Acid-Base Catalyst in the Reaction Catalyzed by Fructose-2,6-bisphosphatase
|
|
7171
|
Glutamate 325 is a General Acid-Base Catalyst in the Reaction Catalyzed by Fructose-2,6-bisphosphatase
|
|
7172
|
Glutamate 325 is a General Acid-Base Catalyst in the Reaction Catalyzed by Fructose-2,6-bisphosphatase
|
|
7173
|
Glutamate 325 is a General Acid-Base Catalyst in the Reaction Catalyzed by Fructose-2,6-bisphosphatase
|
|
7174
|
Glutamate 325 is a General Acid-Base Catalyst in the Reaction Catalyzed by Fructose-2,6-bisphosphatase
|
|
7175
|
Glutamate 325 is a General Acid-Base Catalyst in the Reaction Catalyzed by Fructose-2,6-bisphosphatase
|
|
7176
|
Glutamate 325 is a General Acid-Base Catalyst in the Reaction Catalyzed by Fructose-2,6-bisphosphatase
|
|
7177
|
Glutamate 325 is a General Acid-Base Catalyst in the Reaction Catalyzed by Fructose-2,6-bisphosphatase
|
|
7178
|
Evidence for an essential carboxyl group for yeast phosphoglycerate kinase. Reaction with Woodward`s reagent K
|
|
7179
|
Evidence for an essential carboxyl group for yeast phosphoglycerate kinase. Reaction with Woodward`s reagent K
|
|
7180
|
Evidence for an essential carboxyl group for yeast phosphoglycerate kinase. Reaction with Woodward`s reagent K
|
|
7181
|
Evidence for an essential carboxyl group for yeast phosphoglycerate kinase. Reaction with Woodward`s reagent K
|
|
7182
|
Evidence for an essential carboxyl group for yeast phosphoglycerate kinase. Reaction with Woodward`s reagent K
|
|
7183
|
Sequence analysis and expression of the arginine-deiminase and carbamate-kinase genes of pseudomonas ...
|
|
7184
|
Sequence analysis and expression of the arginine-deiminase and carbamate-kinase genes of pseudomonas ...
|
|
7185
|
Sequence analysis and expression of the arginine-deiminase and carbamate-kinase genes of pseudomonas ...
|
|
7186
|
Sequence analysis and expression of the arginine-deiminase and carbamate-kinase genes of pseudomonas ...
|
|
7187
|
cDNA cloning, expression, and mutagenesis study of liver-type Prostaglandin F Synthase
|
|
7188
|
cDNA cloning, expression, and mutagenesis study of liver-type Prostaglandin F Synthase
|
|
7189
|
cDNA cloning, expression, and mutagenesis study of liver-type Prostaglandin F Synthase
|
|
7190
|
cDNA cloning, expression, and mutagenesis study of liver-type Prostaglandin F Synthase
|
|
7191
|
cDNA cloning, expression, and mutagenesis study of liver-type Prostaglandin F Synthase
|
|
7192
|
cDNA cloning, expression, and mutagenesis study of liver-type Prostaglandin F Synthase
|
|
7193
|
cDNA cloning, expression, and mutagenesis study of liver-type Prostaglandin F Synthase
|
|
7194
|
cDNA cloning, expression, and mutagenesis study of liver-type Prostaglandin F Synthase
|
|
7195
|
cDNA cloning, expression, and mutagenesis study of liver-type Prostaglandin F Synthase
|
|
7196
|
cDNA cloning, expression, and mutagenesis study of liver-type Prostaglandin F Synthase
|
|
7197
|
cDNA cloning, expression, and mutagenesis study of liver-type Prostaglandin F Synthase
|
|
7198
|
cDNA cloning, expression, and mutagenesis study of liver-type Prostaglandin F Synthase
|
|
7199
|
cDNA cloning, expression, and mutagenesis study of liver-type Prostaglandin F Synthase
|
|
7200
|
cDNA cloning, expression, and mutagenesis study of liver-type Prostaglandin F Synthase
|
|
7201
|
cDNA cloning, expression, and mutagenesis study of liver-type Prostaglandin F Synthase
|
|
7202
|
cDNA cloning, expression, and mutagenesis study of liver-type Prostaglandin F Synthase
|
|
7203
|
cDNA cloning, expression, and mutagenesis study of liver-type Prostaglandin F Synthase
|
|
7204
|
cDNA cloning, expression, and mutagenesis study of liver-type Prostaglandin F Synthase
|
|
7205
|
cDNA cloning, expression, and mutagenesis study of liver-type Prostaglandin F Synthase
|
|
7206
|
cDNA cloning, expression, and mutagenesis study of liver-type Prostaglandin F Synthase
|
|
7207
|
cDNA cloning, expression, and mutagenesis study of liver-type Prostaglandin F Synthase
|
|
7208
|
cDNA cloning, expression, and mutagenesis study of liver-type Prostaglandin F Synthase
|
|
7209
|
cDNA cloning, expression, and mutagenesis study of liver-type Prostaglandin F Synthase
|
|
7210
|
cDNA cloning, expression, and mutagenesis study of liver-type Prostaglandin F Synthase
|
|
7211
|
Identification and Significance of Glucokinase in Transplantable Insulinomas
|
|
7212
|
Identification and Significance of Glucokinase in Transplantable Insulinomas
|
|
7213
|
Identification and Significance of Glucokinase in Transplantable Insulinomas
|
|
7214
|
Substitution of the conserved Arg-Tyr dyad selectively disrupts the hydrolysis phase of the IMP dehydrogenase ...
|
|
7215
|
Substitution of the conserved Arg-Tyr dyad selectively disrupts the hydrolysis phase of the IMP dehydrogenase ...
|
|
7216
|
Substitution of the conserved Arg-Tyr dyad selectively disrupts the hydrolysis phase of the IMP dehydrogenase ...
|
|
7217
|
Substitution of the conserved Arg-Tyr dyad selectively disrupts the hydrolysis phase of the IMP dehydrogenase ...
|
|
7218
|
Substitution of the conserved Arg-Tyr dyad selectively disrupts the hydrolysis phase of the IMP dehydrogenase ...
|
|
7219
|
Substitution of the conserved Arg-Tyr dyad selectively disrupts the hydrolysis phase of the IMP dehydrogenase ...
|
|
7220
|
Substitution of the conserved Arg-Tyr dyad selectively disrupts the hydrolysis phase of the IMP dehydrogenase ...
|
|
7221
|
Substitution of the conserved Arg-Tyr dyad selectively disrupts the hydrolysis phase of the IMP dehydrogenase ...
|
|
7222
|
Substitution of the conserved Arg-Tyr dyad selectively disrupts the hydrolysis phase of the IMP dehydrogenase ...
|
|
7223
|
Substitution of the conserved Arg-Tyr dyad selectively disrupts the hydrolysis phase of the IMP dehydrogenase ...
|
|
7224
|
Substitution of the conserved Arg-Tyr dyad selectively disrupts the hydrolysis phase of the IMP dehydrogenase ...
|
|
7225
|
Substitution of the conserved Arg-Tyr dyad selectively disrupts the hydrolysis phase of the IMP dehydrogenase ...
|
|
7226
|
Purification and characterization of glucose-6-phosphate isomerase from Bacillus stearothermophilus
|
|
7227
|
Purification and characterization of glucose-6-phosphate isomerase from Bacillus stearothermophilus
|
|
7228
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7229
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7230
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7231
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7232
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7233
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7234
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7235
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7236
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7237
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7238
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7239
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7240
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7241
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7242
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7243
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7244
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7245
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7246
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7247
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7248
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7249
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7250
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7251
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7252
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7253
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7254
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7255
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7256
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7257
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7258
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7259
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7260
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7261
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7262
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7263
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7264
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7265
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7266
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7267
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7268
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7269
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7270
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7271
|
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human ...
|
|
7272
|
Catalytic, noncatalytic, and inhibitory phenomena: kinetic analysis of (4-hydroxyphenyl)pyruvate dioxygenase ...
|
|
7273
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7274
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7275
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7276
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7277
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7278
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7279
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7280
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7281
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7282
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7283
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7284
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7285
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7286
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7287
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7288
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7289
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7290
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7291
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7292
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7293
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7294
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7295
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7296
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7297
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7298
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7299
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7300
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7301
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7302
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7303
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7304
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7305
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7306
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7307
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7308
|
Steroid-binding site residues dictate optimal substrate positioning in rat 3alpha-hydroxysteroid dehydrogenase ...
|
|
7309
|
Steady-State and Pre-Steady-State Kinetic Analysis of Mycobacterium tuberculosis Pantothenate Synthetase
|
|
7310
|
Steady-State and Pre-Steady-State Kinetic Analysis of Mycobacterium tuberculosis Pantothenate Synthetase
|
|
7311
|
Steady-State and Pre-Steady-State Kinetic Analysis of Mycobacterium tuberculosis Pantothenate Synthetase
|
|
7312
|
Steady-State and Pre-Steady-State Kinetic Analysis of Mycobacterium tuberculosis Pantothenate Synthetase
|
|
7313
|
Steady-State and Pre-Steady-State Kinetic Analysis of Mycobacterium tuberculosis Pantothenate Synthetase
|
|
7314
|
Steady-State and Pre-Steady-State Kinetic Analysis of Mycobacterium tuberculosis Pantothenate Synthetase
|
|
7315
|
Steady-State and Pre-Steady-State Kinetic Analysis of Mycobacterium tuberculosis Pantothenate Synthetase
|
|
7316
|
Steady-State and Pre-Steady-State Kinetic Analysis of Mycobacterium tuberculosis Pantothenate Synthetase
|
|
7317
|
Steady-State and Pre-Steady-State Kinetic Analysis of Mycobacterium tuberculosis Pantothenate Synthetase
|
|
7318
|
Steady-State and Pre-Steady-State Kinetic Analysis of Mycobacterium tuberculosis Pantothenate Synthetase
|
|
7319
|
Steady-State and Pre-Steady-State Kinetic Analysis of Mycobacterium tuberculosis Pantothenate Synthetase
|
|
7320
|
Steady-State and Pre-Steady-State Kinetic Analysis of Mycobacterium tuberculosis Pantothenate Synthetase
|
|
7321
|
Steady-State and Pre-Steady-State Kinetic Analysis of Mycobacterium tuberculosis Pantothenate Synthetase
|
|
7322
|
Steady-State and Pre-Steady-State Kinetic Analysis of Mycobacterium tuberculosis Pantothenate Synthetase
|
|
7323
|
Steady-State and Pre-Steady-State Kinetic Analysis of Mycobacterium tuberculosis Pantothenate Synthetase
|
|
7324
|
Steady-State and Pre-Steady-State Kinetic Analysis of Mycobacterium tuberculosis Pantothenate Synthetase
|
|
7325
|
Steady-State and Pre-Steady-State Kinetic Analysis of Mycobacterium tuberculosis Pantothenate Synthetase
|
|
7326
|
Steady-State and Pre-Steady-State Kinetic Analysis of Mycobacterium tuberculosis Pantothenate Synthetase
|
|
7327
|
A Case for Reverse Protonation: Identification of Glu160 as an Acid/Base Catalyst in Thermoanaerobacterium ...
|
|
7328
|
A Case for Reverse Protonation: Identification of Glu160 as an Acid/Base Catalyst in Thermoanaerobacterium ...
|
|
7329
|
A Case for Reverse Protonation: Identification of Glu160 as an Acid/Base Catalyst in Thermoanaerobacterium ...
|
|
7330
|
A Case for Reverse Protonation: Identification of Glu160 as an Acid/Base Catalyst in Thermoanaerobacterium ...
|
|
7331
|
A Case for Reverse Protonation: Identification of Glu160 as an Acid/Base Catalyst in Thermoanaerobacterium ...
|
|
7332
|
A Case for Reverse Protonation: Identification of Glu160 as an Acid/Base Catalyst in Thermoanaerobacterium ...
|
|
7333
|
A Case for Reverse Protonation: Identification of Glu160 as an Acid/Base Catalyst in Thermoanaerobacterium ...
|
|
7334
|
A Case for Reverse Protonation: Identification of Glu160 as an Acid/Base Catalyst in Thermoanaerobacterium ...
|
|
7335
|
A Case for Reverse Protonation: Identification of Glu160 as an Acid/Base Catalyst in Thermoanaerobacterium ...
|
|
7336
|
A Case for Reverse Protonation: Identification of Glu160 as an Acid/Base Catalyst in Thermoanaerobacterium ...
|
|
7337
|
A Case for Reverse Protonation: Identification of Glu160 as an Acid/Base Catalyst in Thermoanaerobacterium ...
|
|
7338
|
A Case for Reverse Protonation: Identification of Glu160 as an Acid/Base Catalyst in Thermoanaerobacterium ...
|
|
7339
|
A Case for Reverse Protonation: Identification of Glu160 as an Acid/Base Catalyst in Thermoanaerobacterium ...
|
|
7340
|
Kinetic analysis of Pseudomonas aeruginosa arginine deiminase mutants and alternate substrates provides ...
|
|
7341
|
Kinetic analysis of Pseudomonas aeruginosa arginine deiminase mutants and alternate substrates provides ...
|
|
7342
|
Kinetic analysis of Pseudomonas aeruginosa arginine deiminase mutants and alternate substrates provides ...
|
|
7343
|
Kinetic analysis of Pseudomonas aeruginosa arginine deiminase mutants and alternate substrates provides ...
|
|
7344
|
Kinetic analysis of Pseudomonas aeruginosa arginine deiminase mutants and alternate substrates provides ...
|
|
7345
|
Kinetic analysis of Pseudomonas aeruginosa arginine deiminase mutants and alternate substrates provides ...
|
|
7346
|
Kinetic analysis of Pseudomonas aeruginosa arginine deiminase mutants and alternate substrates provides ...
|
|
7347
|
Kinetic analysis of Pseudomonas aeruginosa arginine deiminase mutants and alternate substrates provides ...
|
|
7348
|
Kinetic analysis of Pseudomonas aeruginosa arginine deiminase mutants and alternate substrates provides ...
|
|
7349
|
Kinetic analysis of Pseudomonas aeruginosa arginine deiminase mutants and alternate substrates provides ...
|
|
7350
|
Kinetic analysis of Pseudomonas aeruginosa arginine deiminase mutants and alternate substrates provides ...
|
|
7351
|
Kinetic analysis of Pseudomonas aeruginosa arginine deiminase mutants and alternate substrates provides ...
|
|
7352
|
Kinetic analysis of Pseudomonas aeruginosa arginine deiminase mutants and alternate substrates provides ...
|
|
7353
|
Isolation and characterization of enolase from the rhesus monkey (Macaca mulatta)
|
|
7354
|
Isolation and characterization of enolase from the rhesus monkey (Macaca mulatta)
|
|
7355
|
Isolation and characterization of enolase from the rhesus monkey (Macaca mulatta)
|
|
7356
|
Isolation and characterization of enolase from the rhesus monkey (Macaca mulatta)
|
|
7357
|
The purification and characterization of Escherichia coli enolase
|
|
7358
|
Tryptophanase from Proteus vulgaris: the conformational rearrangement in the active site, induced by the ...
|
|
7359
|
Tryptophanase from Proteus vulgaris: the conformational rearrangement in the active site, induced by the ...
|
|
7360
|
Tryptophanase from Proteus vulgaris: the conformational rearrangement in the active site, induced by the ...
|
|
7361
|
Tryptophanase from Proteus vulgaris: the conformational rearrangement in the active site, induced by the ...
|
|
7362
|
Tryptophanase from Proteus vulgaris: the conformational rearrangement in the active site, induced by the ...
|
|
7363
|
Tryptophanase from Proteus vulgaris: the conformational rearrangement in the active site, induced by the ...
|
|
7364
|
Tryptophanase from Proteus vulgaris: the conformational rearrangement in the active site, induced by the ...
|
|
7365
|
The first archaeal l-aspartate dehydrogenase from the hyperthermophile Archaeoglobus fulgidus: Gene cloning ...
|
|
7366
|
The first archaeal l-aspartate dehydrogenase from the hyperthermophile Archaeoglobus fulgidus: Gene cloning ...
|
|
7367
|
The first archaeal l-aspartate dehydrogenase from the hyperthermophile Archaeoglobus fulgidus: Gene cloning ...
|
|
7368
|
The first archaeal l-aspartate dehydrogenase from the hyperthermophile Archaeoglobus fulgidus: Gene cloning ...
|
|
7369
|
The first archaeal l-aspartate dehydrogenase from the hyperthermophile Archaeoglobus fulgidus: Gene cloning ...
|
|
7370
|
The first archaeal l-aspartate dehydrogenase from the hyperthermophile Archaeoglobus fulgidus: Gene cloning ...
|
|
7371
|
The first archaeal l-aspartate dehydrogenase from the hyperthermophile Archaeoglobus fulgidus: Gene cloning ...
|
|
7372
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7373
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7374
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7375
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7376
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7377
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7378
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7379
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7380
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7381
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7382
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7383
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7384
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7385
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7386
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7387
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7388
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7389
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7390
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7391
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7392
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7393
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7394
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7395
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7396
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7397
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7398
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7399
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7400
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7401
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7402
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7403
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7404
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7405
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7406
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7407
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7408
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7409
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7410
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7411
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7412
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7413
|
Molecular interpretation of inhibition by excess substrate in flavocytochrome b2: a study with wild-type and ...
|
|
7414
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7415
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7416
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7417
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7418
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7419
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7420
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7421
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7422
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7423
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7424
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7425
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7426
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7427
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7428
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7429
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7430
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7431
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7432
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7433
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7434
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7435
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7436
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7437
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7438
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7439
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7440
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7441
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7442
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7443
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7444
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7445
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7446
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7447
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7448
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7449
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7450
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7451
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7452
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7453
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7454
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7455
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7456
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7457
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7458
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7459
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7460
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7461
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7462
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7463
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7464
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7465
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7466
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7467
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7468
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7469
|
The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation ...
|
|
7470
|
Purification and properties of arginase from human liver and erythrocytes
|
|
7471
|
Purification and properties of arginase from human liver and erythrocytes
|
|
7472
|
Purification and properties of arginase from human liver and erythrocytes
|
|
7473
|
Purification and properties of arginase from human liver and erythrocytes
|
|
7474
|
The purification and characterization of arginase from saccharomyces cerevisiae
|
|
7475
|
Purification and characterization of the class-II D-fructose 1,6-bisphosphate aldolase from Escherichia coli ...
|
|
7476
|
Aspartate dehydrogenase, a novel enzyme identified from structural and functional studies of TM1643
|
|
7477
|
Aspartate dehydrogenase, a novel enzyme identified from structural and functional studies of TM1643
|
|
7478
|
Aspartate dehydrogenase, a novel enzyme identified from structural and functional studies of TM1643
|
|
7479
|
Aspartate dehydrogenase, a novel enzyme identified from structural and functional studies of TM1643
|
|
7480
|
Aspartate dehydrogenase, a novel enzyme identified from structural and functional studies of TM1643
|
|
7481
|
Aspartate dehydrogenase, a novel enzyme identified from structural and functional studies of TM1643
|
|
7482
|
Calorimetric determination of thermodynamic parameters of reaction reveals different enthalpic compensations ...
|
|
7483
|
Calorimetric determination of thermodynamic parameters of reaction reveals different enthalpic compensations ...
|
|
7484
|
Characterisation of yeast phosphoglycerate kinase modified by mutagenesis at residue 21
|
|
7485
|
Characterisation of yeast phosphoglycerate kinase modified by mutagenesis at residue 21
|
|
7486
|
Characterisation of yeast phosphoglycerate kinase modified by mutagenesis at residue 21
|
|
7487
|
Characterisation of yeast phosphoglycerate kinase modified by mutagenesis at residue 21
|
|
7488
|
Characterisation of yeast phosphoglycerate kinase modified by mutagenesis at residue 21
|
|
7489
|
Characterisation of yeast phosphoglycerate kinase modified by mutagenesis at residue 21
|
|
7490
|
Periplasmic phosphatases in salmonella typhimurium LT2
|
|
7491
|
Periplasmic phosphatases in salmonella typhimurium LT2
|
|
7492
|
Polyamine-deficient neurospora crassa mutants and synthesis of cadaverine
|
|
7493
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7494
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7495
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7496
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7497
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7498
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7499
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7500
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7501
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7502
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7503
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7504
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7505
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7506
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7507
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7508
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7509
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7510
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7511
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7512
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7513
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7514
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7515
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7516
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7517
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7518
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7519
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7520
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7521
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7522
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7523
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7524
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7525
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7526
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7527
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7528
|
The effects of temperature and pH on the kinetic properties of heart muscle lactate dehydrogenase from anuran ...
|
|
7529
|
Role of metal ions in catalysis by enolase: an ordered kinetic mechanism for a single substrate enzyme
|
|
7530
|
Role of metal ions in catalysis by enolase: an ordered kinetic mechanism for a single substrate enzyme
|
|
7531
|
Role of metal ions in catalysis by enolase: an ordered kinetic mechanism for a single substrate enzyme
|
|
7532
|
Role of metal ions in catalysis by enolase: an ordered kinetic mechanism for a single substrate enzyme
|
|
7533
|
Role of metal ions in catalysis by enolase: an ordered kinetic mechanism for a single substrate enzyme
|
|
7534
|
Chemical mechanism of the endogenous argininosuccinate lyase activity of duck lens delta2-crystallin
|
|
7535
|
Chemical mechanism of the endogenous argininosuccinate lyase activity of duck lens delta2-crystallin
|
|
7536
|
Assay and kinetics of arginase
|
|
7537
|
Subcellular localization and kinetic properties of arginase from the liver of genypterus maculatus
|
|
7538
|
Subcellular localization and kinetic properties of arginase from the liver of genypterus maculatus
|
|
7539
|
Subcellular localization and kinetic properties of arginase from the liver of genypterus maculatus
|
|
7540
|
Subcellular localization and kinetic properties of arginase from the liver of genypterus maculatus
|
|
7541
|
Metabolic stability of alpha-methylated polyamine derivatives and their use as substitutes for the natural ...
|
|
7542
|
Metabolic stability of alpha-methylated polyamine derivatives and their use as substitutes for the natural ...
|
|
7543
|
Metabolic stability of alpha-methylated polyamine derivatives and their use as substitutes for the natural ...
|
|
7544
|
Metabolic stability of alpha-methylated polyamine derivatives and their use as substitutes for the natural ...
|
|
7545
|
Metabolic stability of alpha-methylated polyamine derivatives and their use as substitutes for the natural ...
|
|
7546
|
Metabolic stability of alpha-methylated polyamine derivatives and their use as substitutes for the natural ...
|
|
7547
|
Metabolic stability of alpha-methylated polyamine derivatives and their use as substitutes for the natural ...
|
|
7548
|
Metabolic stability of alpha-methylated polyamine derivatives and their use as substitutes for the natural ...
|
|
7549
|
Metabolic stability of alpha-methylated polyamine derivatives and their use as substitutes for the natural ...
|
|
7550
|
Metabolic stability of alpha-methylated polyamine derivatives and their use as substitutes for the natural ...
|
|
7551
|
Metabolic stability of alpha-methylated polyamine derivatives and their use as substitutes for the natural ...
|
|
7552
|
Metabolic stability of alpha-methylated polyamine derivatives and their use as substitutes for the natural ...
|
|
7553
|
Metabolic stability of alpha-methylated polyamine derivatives and their use as substitutes for the natural ...
|
|
7554
|
Metabolic stability of alpha-methylated polyamine derivatives and their use as substitutes for the natural ...
|
|
7555
|
Metabolic stability of alpha-methylated polyamine derivatives and their use as substitutes for the natural ...
|
|
7556
|
Metabolic stability of alpha-methylated polyamine derivatives and their use as substitutes for the natural ...
|
|
7557
|
Metabolic stability of alpha-methylated polyamine derivatives and their use as substitutes for the natural ...
|
|
7558
|
Metabolic stability of alpha-methylated polyamine derivatives and their use as substitutes for the natural ...
|
|
7559
|
Metabolic stability of alpha-methylated polyamine derivatives and their use as substitutes for the natural ...
|
|
7560
|
Metabolic stability of alpha-methylated polyamine derivatives and their use as substitutes for the natural ...
|
|
7561
|
Metabolic stability of alpha-methylated polyamine derivatives and their use as substitutes for the natural ...
|
|
7562
|
Physicochemical and catalytic properties of thermostable malate dehydrogenase from an extreme thermophile ...
|
|
7563
|
Physicochemical and catalytic properties of thermostable malate dehydrogenase from an extreme thermophile ...
|
|
7564
|
Physicochemical and catalytic properties of thermostable malate dehydrogenase from an extreme thermophile ...
|
|
7565
|
Physicochemical and catalytic properties of thermostable malate dehydrogenase from an extreme thermophile ...
|
|
7566
|
Physicochemical and catalytic properties of thermostable malate dehydrogenase from an extreme thermophile ...
|
|
7567
|
Physicochemical and catalytic properties of thermostable malate dehydrogenase from an extreme thermophile ...
|
|
7568
|
Physicochemical and catalytic properties of thermostable malate dehydrogenase from an extreme thermophile ...
|
|
7569
|
Physicochemical and catalytic properties of thermostable malate dehydrogenase from an extreme thermophile ...
|
|
7570
|
The mechanism of kynurenine hydrolysis catalyzed by kynureninase
|
|
7571
|
The mechanism of kynurenine hydrolysis catalyzed by kynureninase
|
|
7572
|
The mechanism of kynurenine hydrolysis catalyzed by kynureninase
|
|
7573
|
The mechanism of kynurenine hydrolysis catalyzed by kynureninase
|
|
7574
|
The mechanism of kynurenine hydrolysis catalyzed by kynureninase
|
|
7575
|
The mechanism of kynurenine hydrolysis catalyzed by kynureninase
|
|
7576
|
The mechanism of kynurenine hydrolysis catalyzed by kynureninase
|
|
7577
|
The mechanism of kynurenine hydrolysis catalyzed by kynureninase
|
|
7578
|
The mechanism of kynurenine hydrolysis catalyzed by kynureninase
|
|
7579
|
Enzymatic and biochemical properties of a novel human serine dehydratase isoform
|
|
7580
|
Enzymatic and biochemical properties of a novel human serine dehydratase isoform
|
|
7581
|
Enzymatic and biochemical properties of a novel human serine dehydratase isoform
|
|
7582
|
Enzymatic and biochemical properties of a novel human serine dehydratase isoform
|
|
7583
|
Enzymatic and biochemical properties of a novel human serine dehydratase isoform
|
|
7584
|
Enzymatic and biochemical properties of a novel human serine dehydratase isoform
|
|
7585
|
Enzymatic and biochemical properties of a novel human serine dehydratase isoform
|
|
7586
|
Enzymatic and biochemical properties of a novel human serine dehydratase isoform
|
|
7587
|
Enzymatic and biochemical properties of a novel human serine dehydratase isoform
|
|
7588
|
Enzymatic and biochemical properties of a novel human serine dehydratase isoform
|
|
7589
|
Enzymatic and biochemical properties of a novel human serine dehydratase isoform
|
|
7590
|
Enzymatic and biochemical properties of a novel human serine dehydratase isoform
|
|
7591
|
Characterisation of arginase from the extreme thermophile bacillus caldovelox
|
|
7592
|
Characterisation of arginase from the extreme thermophile bacillus caldovelox
|
|
7593
|
Characterisation of arginase from the extreme thermophile bacillus caldovelox
|
|
7594
|
Characterisation of arginase from the extreme thermophile bacillus caldovelox
|
|
7595
|
Role of His505 in the soluble fumarate reductase from Shewanella frigidimarina
|
|
7596
|
Role of His505 in the soluble fumarate reductase from Shewanella frigidimarina
|
|
7597
|
Role of His505 in the soluble fumarate reductase from Shewanella frigidimarina
|
|
7598
|
Role of His505 in the soluble fumarate reductase from Shewanella frigidimarina
|
|
7599
|
Role of His505 in the soluble fumarate reductase from Shewanella frigidimarina
|
|
7600
|
Role of His505 in the soluble fumarate reductase from Shewanella frigidimarina
|
|
7601
|
Role of His505 in the soluble fumarate reductase from Shewanella frigidimarina
|
|
7602
|
Role of His505 in the soluble fumarate reductase from Shewanella frigidimarina
|
|
7603
|
His68 and His141 are critical contributors to the intersubunit catalytic site of adenylosuccinate lyase of ...
|
|
7604
|
His68 and His141 are critical contributors to the intersubunit catalytic site of adenylosuccinate lyase of ...
|
|
7605
|
His68 and His141 are critical contributors to the intersubunit catalytic site of adenylosuccinate lyase of ...
|
|
7606
|
His68 and His141 are critical contributors to the intersubunit catalytic site of adenylosuccinate lyase of ...
|
|
7607
|
His68 and His141 are critical contributors to the intersubunit catalytic site of adenylosuccinate lyase of ...
|
|
7608
|
His68 and His141 are critical contributors to the intersubunit catalytic site of adenylosuccinate lyase of ...
|
|
7609
|
His68 and His141 are critical contributors to the intersubunit catalytic site of adenylosuccinate lyase of ...
|
|
7610
|
His68 and His141 are critical contributors to the intersubunit catalytic site of adenylosuccinate lyase of ...
|
|
7611
|
His68 and His141 are critical contributors to the intersubunit catalytic site of adenylosuccinate lyase of ...
|
|
7612
|
His68 and His141 are critical contributors to the intersubunit catalytic site of adenylosuccinate lyase of ...
|
|
7613
|
His68 and His141 are critical contributors to the intersubunit catalytic site of adenylosuccinate lyase of ...
|
|
7614
|
His68 and His141 are critical contributors to the intersubunit catalytic site of adenylosuccinate lyase of ...
|
|
7615
|
His68 and His141 are critical contributors to the intersubunit catalytic site of adenylosuccinate lyase of ...
|
|
7616
|
His68 and His141 are critical contributors to the intersubunit catalytic site of adenylosuccinate lyase of ...
|
|
7617
|
His68 and His141 are critical contributors to the intersubunit catalytic site of adenylosuccinate lyase of ...
|
|
7618
|
His68 and His141 are critical contributors to the intersubunit catalytic site of adenylosuccinate lyase of ...
|
|
7619
|
His68 and His141 are critical contributors to the intersubunit catalytic site of adenylosuccinate lyase of ...
|
|
7620
|
Redox properties of flavocytochrome c3 from Shewanella frigidimarina NCIMB400
|
|
7621
|
Redox properties of flavocytochrome c3 from Shewanella frigidimarina NCIMB400
|
|
7622
|
Redox properties of flavocytochrome c3 from Shewanella frigidimarina NCIMB400
|
|
7623
|
Redox properties of flavocytochrome c3 from Shewanella frigidimarina NCIMB400
|
|
7624
|
Redox properties of flavocytochrome c3 from Shewanella frigidimarina NCIMB400
|
|
7625
|
Redox properties of flavocytochrome c3 from Shewanella frigidimarina NCIMB400
|
|
7626
|
Redox properties of flavocytochrome c3 from Shewanella frigidimarina NCIMB400
|
|
7627
|
Redox properties of flavocytochrome c3 from Shewanella frigidimarina NCIMB400
|
|
7628
|
Redox properties of flavocytochrome c3 from Shewanella frigidimarina NCIMB400
|
|
7629
|
Redox properties of flavocytochrome c3 from Shewanella frigidimarina NCIMB400
|
|
7630
|
Redox properties of flavocytochrome c3 from Shewanella frigidimarina NCIMB400
|
|
7631
|
Identification of the active site acid/base catalyst in a bacterial fumarate reductase: a kinetic and ...
|
|
7632
|
Identification of the active site acid/base catalyst in a bacterial fumarate reductase: a kinetic and ...
|
|
7633
|
Identification of the active site acid/base catalyst in a bacterial fumarate reductase: a kinetic and ...
|
|
7634
|
Identification of the active site acid/base catalyst in a bacterial fumarate reductase: a kinetic and ...
|
|
7635
|
Identification of the active site acid/base catalyst in a bacterial fumarate reductase: a kinetic and ...
|
|
7636
|
Identification of the active site acid/base catalyst in a bacterial fumarate reductase: a kinetic and ...
|
|
7637
|
Identification of the active site acid/base catalyst in a bacterial fumarate reductase: a kinetic and ...
|
|
7638
|
Identification of the active site acid/base catalyst in a bacterial fumarate reductase: a kinetic and ...
|
|
7639
|
Identification of the active site acid/base catalyst in a bacterial fumarate reductase: a kinetic and ...
|
|
7640
|
Identification of the active site acid/base catalyst in a bacterial fumarate reductase: a kinetic and ...
|
|
7641
|
Identification of the active site acid/base catalyst in a bacterial fumarate reductase: a kinetic and ...
|
|
7642
|
Identification of the active site acid/base catalyst in a bacterial fumarate reductase: a kinetic and ...
|
|
7643
|
Identification of the active site acid/base catalyst in a bacterial fumarate reductase: a kinetic and ...
|
|
7644
|
Identification of the active site acid/base catalyst in a bacterial fumarate reductase: a kinetic and ...
|
|
7645
|
Arginase from human full-term placenta
|
|
7646
|
Arginase from human full-term placenta
|
|
7647
|
Arginase from human full-term placenta
|
|
7648
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7649
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7650
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7651
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7652
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7653
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7654
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7655
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7656
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7657
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7658
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7659
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7660
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7661
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7662
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7663
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7664
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7665
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7666
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7667
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7668
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7669
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7670
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7671
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7672
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7673
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7674
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7675
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7676
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7677
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7678
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7679
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7680
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7681
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7682
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7683
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7684
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7685
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7686
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7687
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7688
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7689
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7690
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7691
|
Engineering the substrate specificity of porcine kidney D-amino acid oxidase by mutagenesis of the ...
|
|
7692
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7693
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7694
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7695
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7696
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7697
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7698
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7699
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7700
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7701
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7702
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7703
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7704
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7705
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7706
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7707
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7708
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7709
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7710
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7711
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7712
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7713
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7714
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7715
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7716
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7717
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7718
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7719
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7720
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7721
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7722
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7723
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7724
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7725
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7726
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7727
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7728
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7729
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7730
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7731
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7732
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7733
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7734
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7735
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7736
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7737
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7738
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7739
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7740
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7741
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7742
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7743
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7744
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7745
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7746
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7747
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7748
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7749
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7750
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7751
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7752
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7753
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7754
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7755
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7756
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7757
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7758
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7759
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7760
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7761
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7762
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7763
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7764
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7765
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7766
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7767
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7768
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7769
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7770
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7771
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7772
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7773
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7774
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7775
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7776
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7777
|
Single amino acid substitution in Bacillus sphaericus phenylalanine dehydrogenase dramatically increases its ...
|
|
7778
|
Effect of phosphoglucose isomerase and glucose 6-phosphate on UDP-N-acetylglucosamine inhibition of ...
|
|
7779
|
Effect of phosphoglucose isomerase and glucose 6-phosphate on UDP-N-acetylglucosamine inhibition of ...
|
|
7780
|
The active site and substrate-binding mode of 1-aminocyclopropane-1-carboxylate oxidase determined by ...
|
|
7781
|
The active site and substrate-binding mode of 1-aminocyclopropane-1-carboxylate oxidase determined by ...
|
|
7782
|
The active site and substrate-binding mode of 1-aminocyclopropane-1-carboxylate oxidase determined by ...
|
|
7783
|
The active site and substrate-binding mode of 1-aminocyclopropane-1-carboxylate oxidase determined by ...
|
|
7784
|
The active site and substrate-binding mode of 1-aminocyclopropane-1-carboxylate oxidase determined by ...
|
|
7785
|
The active site and substrate-binding mode of 1-aminocyclopropane-1-carboxylate oxidase determined by ...
|
|
7786
|
The active site and substrate-binding mode of 1-aminocyclopropane-1-carboxylate oxidase determined by ...
|
|
7787
|
The active site and substrate-binding mode of 1-aminocyclopropane-1-carboxylate oxidase determined by ...
|
|
7788
|
The active site and substrate-binding mode of 1-aminocyclopropane-1-carboxylate oxidase determined by ...
|
|
7789
|
The active site and substrate-binding mode of 1-aminocyclopropane-1-carboxylate oxidase determined by ...
|
|
7790
|
The active site and substrate-binding mode of 1-aminocyclopropane-1-carboxylate oxidase determined by ...
|
|
7791
|
The active site and substrate-binding mode of 1-aminocyclopropane-1-carboxylate oxidase determined by ...
|
|
7792
|
The active site and substrate-binding mode of 1-aminocyclopropane-1-carboxylate oxidase determined by ...
|
|
7793
|
The active site and substrate-binding mode of 1-aminocyclopropane-1-carboxylate oxidase determined by ...
|
|
7794
|
The active site and substrate-binding mode of 1-aminocyclopropane-1-carboxylate oxidase determined by ...
|
|
7795
|
The active site and substrate-binding mode of 1-aminocyclopropane-1-carboxylate oxidase determined by ...
|
|
7796
|
Interpretation of the kinetics of consecutive enzyme-catalyzed reactions
|
|
7797
|
Interpretation of the kinetics of consecutive enzyme-catalyzed reactions
|
|
7798
|
Interpretation of the kinetics of consecutive enzyme-catalyzed reactions
|
|
7799
|
Interpretation of the kinetics of consecutive enzyme-catalyzed reactions
|
|
7800
|
Purification and characterization of arginase from neurospora crassa
|
|
7801
|
Cross-talk between thiamin diphosphate binding and phosphorylation loop conformation in human branched-chain ...
|
|
7802
|
Cross-talk between thiamin diphosphate binding and phosphorylation loop conformation in human branched-chain ...
|
|
7803
|
Cross-talk between thiamin diphosphate binding and phosphorylation loop conformation in human branched-chain ...
|
|
7804
|
Cross-talk between thiamin diphosphate binding and phosphorylation loop conformation in human branched-chain ...
|
|
7805
|
Cross-talk between thiamin diphosphate binding and phosphorylation loop conformation in human branched-chain ...
|
|
7806
|
Cross-talk between thiamin diphosphate binding and phosphorylation loop conformation in human branched-chain ...
|
|
7807
|
Cross-talk between thiamin diphosphate binding and phosphorylation loop conformation in human branched-chain ...
|
|
7808
|
Cross-talk between thiamin diphosphate binding and phosphorylation loop conformation in human branched-chain ...
|
|
7809
|
Cross-talk between thiamin diphosphate binding and phosphorylation loop conformation in human branched-chain ...
|
|
7810
|
Cross-talk between thiamin diphosphate binding and phosphorylation loop conformation in human branched-chain ...
|
|
7811
|
Arginase in lactating bovine mammary glands: implications in proline synthesis
|
|
7812
|
Arginase in lactating bovine mammary glands: implications in proline synthesis
|
|
7813
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7814
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7815
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7816
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7817
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7818
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7819
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7820
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7821
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7822
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7823
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7824
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7825
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7826
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7827
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7828
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7829
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7830
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7831
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7832
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7833
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7834
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7835
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7836
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7837
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7838
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7839
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7840
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7841
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7842
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7843
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7844
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7845
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7846
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7847
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7848
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7849
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7850
|
Delineation of the Roles of Amino Acids Involved in the Catalytic Functions of Leuconostoc mesenteroides ...
|
|
7851
|
Lysine-21 of Leuconostoc mesenteroides glucose 6-phosphate dehydrogenase participates in substrate binding ...
|
|
7852
|
Lysine-21 of Leuconostoc mesenteroides glucose 6-phosphate dehydrogenase participates in substrate binding ...
|
|
7853
|
Lysine-21 of Leuconostoc mesenteroides glucose 6-phosphate dehydrogenase participates in substrate binding ...
|
|
7854
|
Lysine-21 of Leuconostoc mesenteroides glucose 6-phosphate dehydrogenase participates in substrate binding ...
|
|
7855
|
Lysine-21 of Leuconostoc mesenteroides glucose 6-phosphate dehydrogenase participates in substrate binding ...
|
|
7856
|
Lysine-21 of Leuconostoc mesenteroides glucose 6-phosphate dehydrogenase participates in substrate binding ...
|
|
7857
|
Arginyl-tRNA synthetase from escherichia coli influence of arginine biosynthetic precursors on the charging ...
|
|
7858
|
Arginyl-tRNA synthetase from escherichia coli influence of arginine biosynthetic precursors on the charging ...
|
|
7859
|
Arginyl-tRNA synthetase from escherichia coli influence of arginine biosynthetic precursors on the charging ...
|
|
7860
|
Arginyl-tRNA synthetase from escherichia coli influence of arginine biosynthetic precursors on the charging ...
|
|
7861
|
Arginyl-tRNA synthetase from escherichia coli influence of arginine biosynthetic precursors on the charging ...
|
|
7862
|
Arginyl-tRNA synthetase from escherichia coli influence of arginine biosynthetic precursors on the charging ...
|
|
7863
|
Arginyl-tRNA synthetase from escherichia coli influence of arginine biosynthetic precursors on the charging ...
|
|
7864
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7865
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7866
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7867
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7868
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7869
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7870
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7871
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7872
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7873
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7874
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7875
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7876
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7877
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7878
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7879
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7880
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7881
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7882
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7883
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7884
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7885
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7886
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7887
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7888
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7889
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7890
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7891
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7892
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7893
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7894
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7895
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7896
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7897
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7898
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7899
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7900
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7901
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7902
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7903
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7904
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7905
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7906
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7907
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7908
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7909
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7910
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7911
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7912
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
7913
|
Ser95, Asn97, and Thr78 are important for the catalytic function of porcine NADP-dependent isocitrate ...
|
|
7914
|
Ser95, Asn97, and Thr78 are important for the catalytic function of porcine NADP-dependent isocitrate ...
|
|
7915
|
Ser95, Asn97, and Thr78 are important for the catalytic function of porcine NADP-dependent isocitrate ...
|
|
7916
|
Ser95, Asn97, and Thr78 are important for the catalytic function of porcine NADP-dependent isocitrate ...
|
|
7917
|
Ser95, Asn97, and Thr78 are important for the catalytic function of porcine NADP-dependent isocitrate ...
|
|
7918
|
Ser95, Asn97, and Thr78 are important for the catalytic function of porcine NADP-dependent isocitrate ...
|
|
7919
|
Ser95, Asn97, and Thr78 are important for the catalytic function of porcine NADP-dependent isocitrate ...
|
|
7920
|
Ser95, Asn97, and Thr78 are important for the catalytic function of porcine NADP-dependent isocitrate ...
|
|
7921
|
Ser95, Asn97, and Thr78 are important for the catalytic function of porcine NADP-dependent isocitrate ...
|
|
7922
|
Ser95, Asn97, and Thr78 are important for the catalytic function of porcine NADP-dependent isocitrate ...
|
|
7923
|
Ser95, Asn97, and Thr78 are important for the catalytic function of porcine NADP-dependent isocitrate ...
|
|
7924
|
Ser95, Asn97, and Thr78 are important for the catalytic function of porcine NADP-dependent isocitrate ...
|
|
7925
|
Ser95, Asn97, and Thr78 are important for the catalytic function of porcine NADP-dependent isocitrate ...
|
|
7926
|
Ser95, Asn97, and Thr78 are important for the catalytic function of porcine NADP-dependent isocitrate ...
|
|
7927
|
Ser95, Asn97, and Thr78 are important for the catalytic function of porcine NADP-dependent isocitrate ...
|
|
7928
|
Ser95, Asn97, and Thr78 are important for the catalytic function of porcine NADP-dependent isocitrate ...
|
|
7929
|
Ser95, Asn97, and Thr78 are important for the catalytic function of porcine NADP-dependent isocitrate ...
|
|
7930
|
Ser95, Asn97, and Thr78 are important for the catalytic function of porcine NADP-dependent isocitrate ...
|
|
7931
|
Ser95, Asn97, and Thr78 are important for the catalytic function of porcine NADP-dependent isocitrate ...
|
|
7932
|
Ser95, Asn97, and Thr78 are important for the catalytic function of porcine NADP-dependent isocitrate ...
|
|
7933
|
Ser95, Asn97, and Thr78 are important for the catalytic function of porcine NADP-dependent isocitrate ...
|
|
7934
|
Ser95, Asn97, and Thr78 are important for the catalytic function of porcine NADP-dependent isocitrate ...
|
|
7935
|
Ser95, Asn97, and Thr78 are important for the catalytic function of porcine NADP-dependent isocitrate ...
|
|
7936
|
Ser95, Asn97, and Thr78 are important for the catalytic function of porcine NADP-dependent isocitrate ...
|
|
7937
|
Ser95, Asn97, and Thr78 are important for the catalytic function of porcine NADP-dependent isocitrate ...
|
|
7938
|
Evaluation by mutagenesis of the roles of His309, His315, and His319 in the coenzyme site of pig heart ...
|
|
7939
|
Evaluation by mutagenesis of the roles of His309, His315, and His319 in the coenzyme site of pig heart ...
|
|
7940
|
Evaluation by mutagenesis of the roles of His309, His315, and His319 in the coenzyme site of pig heart ...
|
|
7941
|
Evaluation by mutagenesis of the roles of His309, His315, and His319 in the coenzyme site of pig heart ...
|
|
7942
|
Evaluation by mutagenesis of the roles of His309, His315, and His319 in the coenzyme site of pig heart ...
|
|
7943
|
Evaluation by mutagenesis of the roles of His309, His315, and His319 in the coenzyme site of pig heart ...
|
|
7944
|
Evaluation by mutagenesis of the roles of His309, His315, and His319 in the coenzyme site of pig heart ...
|
|
7945
|
Evaluation by mutagenesis of the roles of His309, His315, and His319 in the coenzyme site of pig heart ...
|
|
7946
|
Evaluation by mutagenesis of the roles of His309, His315, and His319 in the coenzyme site of pig heart ...
|
|
7947
|
Evaluation by mutagenesis of the roles of His309, His315, and His319 in the coenzyme site of pig heart ...
|
|
7948
|
Evaluation by mutagenesis of the roles of His309, His315, and His319 in the coenzyme site of pig heart ...
|
|
7949
|
Evaluation by mutagenesis of the roles of His309, His315, and His319 in the coenzyme site of pig heart ...
|
|
7950
|
Evaluation by mutagenesis of the roles of His309, His315, and His319 in the coenzyme site of pig heart ...
|
|
7951
|
Evaluation by mutagenesis of the roles of His309, His315, and His319 in the coenzyme site of pig heart ...
|
|
7952
|
Implication by site-directed mutagenesis of Arg314 and Tyr316 in the coenzyme site of pig mitochondrial ...
|
|
7953
|
Implication by site-directed mutagenesis of Arg314 and Tyr316 in the coenzyme site of pig mitochondrial ...
|
|
7954
|
Implication by site-directed mutagenesis of Arg314 and Tyr316 in the coenzyme site of pig mitochondrial ...
|
|
7955
|
Implication by site-directed mutagenesis of Arg314 and Tyr316 in the coenzyme site of pig mitochondrial ...
|
|
7956
|
Implication by site-directed mutagenesis of Arg314 and Tyr316 in the coenzyme site of pig mitochondrial ...
|
|
7957
|
Implication by site-directed mutagenesis of Arg314 and Tyr316 in the coenzyme site of pig mitochondrial ...
|
|
7958
|
Implication by site-directed mutagenesis of Arg314 and Tyr316 in the coenzyme site of pig mitochondrial ...
|
|
7959
|
Implication by site-directed mutagenesis of Arg314 and Tyr316 in the coenzyme site of pig mitochondrial ...
|
|
7960
|
Implication by site-directed mutagenesis of Arg314 and Tyr316 in the coenzyme site of pig mitochondrial ...
|
|
7961
|
Implication by site-directed mutagenesis of Arg314 and Tyr316 in the coenzyme site of pig mitochondrial ...
|
|
7962
|
Implication by site-directed mutagenesis of Arg314 and Tyr316 in the coenzyme site of pig mitochondrial ...
|
|
7963
|
Implication by site-directed mutagenesis of Arg314 and Tyr316 in the coenzyme site of pig mitochondrial ...
|
|
7964
|
Implication by site-directed mutagenesis of Arg314 and Tyr316 in the coenzyme site of pig mitochondrial ...
|
|
7965
|
Implication by site-directed mutagenesis of Arg314 and Tyr316 in the coenzyme site of pig mitochondrial ...
|
|
7966
|
Implication by site-directed mutagenesis of Arg314 and Tyr316 in the coenzyme site of pig mitochondrial ...
|
|
7967
|
Implication by site-directed mutagenesis of Arg314 and Tyr316 in the coenzyme site of pig mitochondrial ...
|
|
7968
|
Implication by site-directed mutagenesis of Arg314 and Tyr316 in the coenzyme site of pig mitochondrial ...
|
|
7969
|
Implication by site-directed mutagenesis of Arg314 and Tyr316 in the coenzyme site of pig mitochondrial ...
|
|
7970
|
Implication by site-directed mutagenesis of Arg314 and Tyr316 in the coenzyme site of pig mitochondrial ...
|
|
7971
|
Implication by site-directed mutagenesis of Arg314 and Tyr316 in the coenzyme site of pig mitochondrial ...
|
|
7972
|
Implication by site-directed mutagenesis of Arg314 and Tyr316 in the coenzyme site of pig mitochondrial ...
|
|
7973
|
Implication by site-directed mutagenesis of Arg314 and Tyr316 in the coenzyme site of pig mitochondrial ...
|
|
7974
|
Implication by site-directed mutagenesis of Arg314 and Tyr316 in the coenzyme site of pig mitochondrial ...
|
|
7975
|
Implication by site-directed mutagenesis of Arg314 and Tyr316 in the coenzyme site of pig mitochondrial ...
|
|
7976
|
Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase
|
|
7977
|
Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase
|
|
7978
|
Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase
|
|
7979
|
Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase
|
|
7980
|
Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase
|
|
7981
|
Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase
|
|
7982
|
Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase
|
|
7983
|
Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase
|
|
7984
|
Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase
|
|
7985
|
Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase
|
|
7986
|
Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase
|
|
7987
|
Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase
|
|
7988
|
Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase
|
|
7989
|
Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase
|
|
7990
|
Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase
|
|
7991
|
Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase
|
|
7992
|
Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase
|
|
7993
|
Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase
|
|
7994
|
Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase
|
|
7995
|
Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase
|
|
7996
|
Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase
|
|
7997
|
Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase
|
|
7998
|
Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase
|
|
7999
|
Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase
|
|
8000
|
Site-directed mutagenesis of essential residues involved in the mechanism of bacterial glycosylasparaginase
|





 If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.
If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.