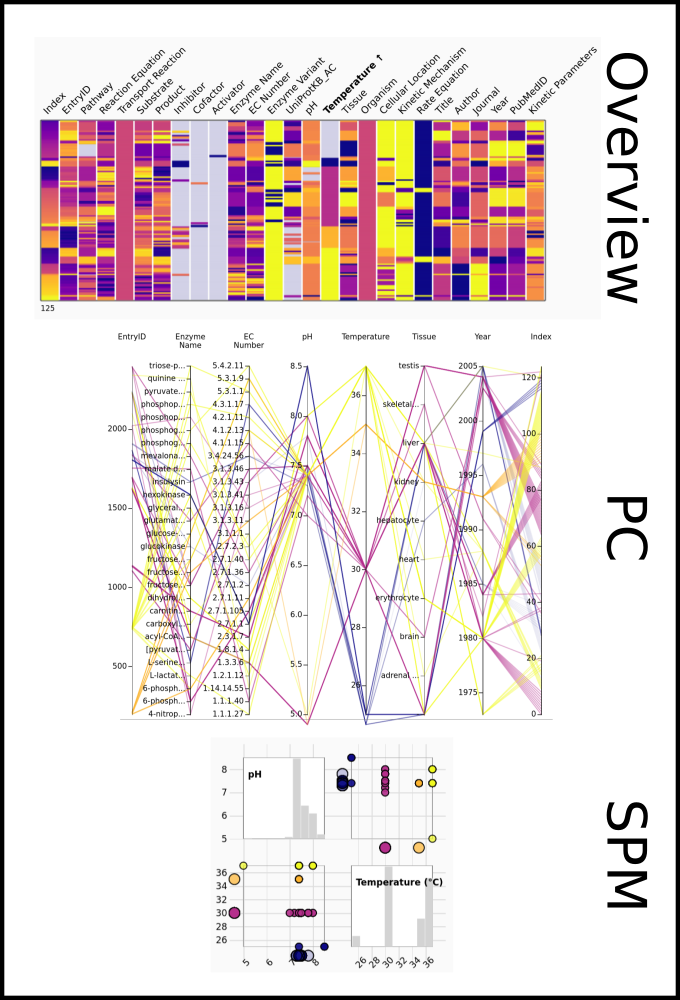
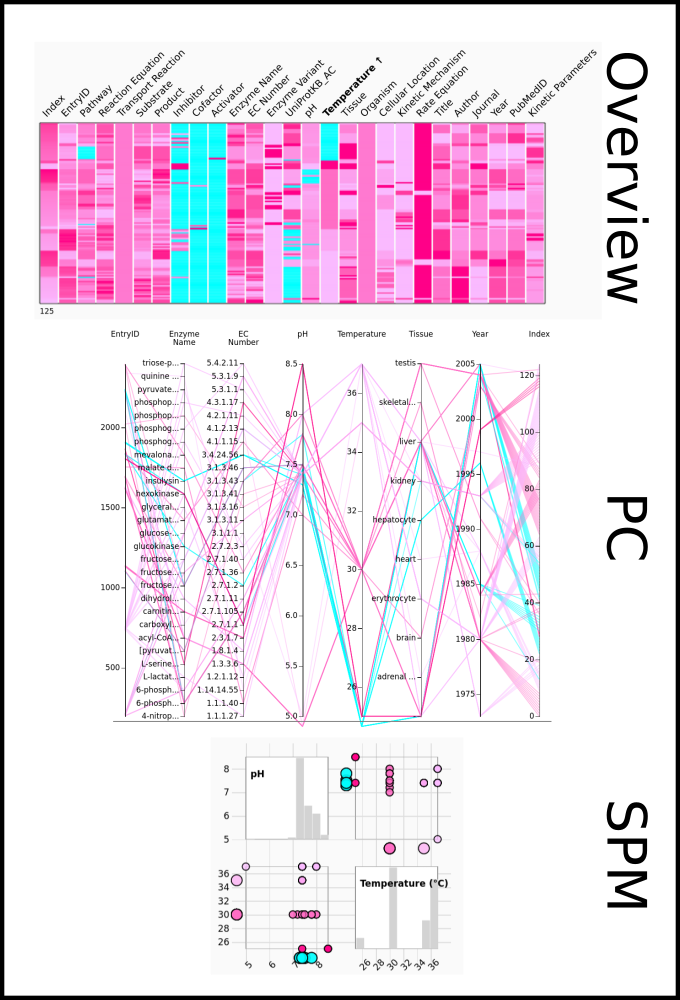
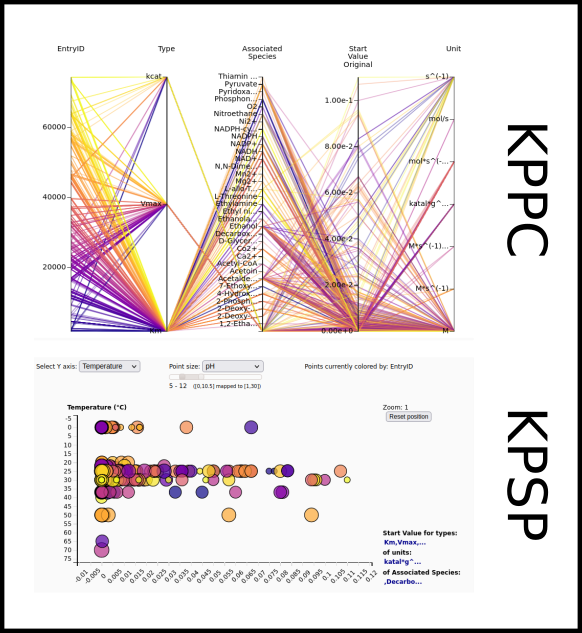
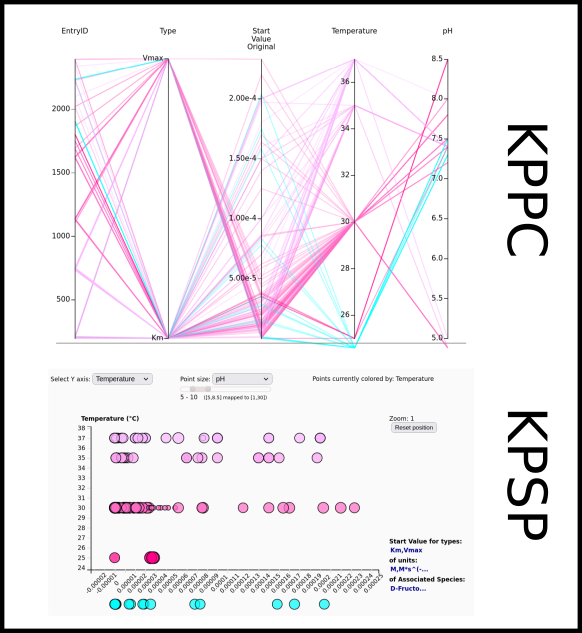
To improve SABIO-RK data interoperability, semantic markup was added to web pages as described and defined by
This structured information makes it easier to discover, collate and analyse our data.
|
70001
|
Multicopper oxidase of Acinetobacter baumannii: Assessing its role in metal homeostasis, stress management and ...
|
|
70002
|
Multicopper oxidase of Acinetobacter baumannii: Assessing its role in metal homeostasis, stress management and ...
|
|
70003
|
Oxidation of phenolate siderophores by the multicopper oxidase encoded by the Escherichia coli yacK gene
|
|
70004
|
Oxidation of phenolate siderophores by the multicopper oxidase encoded by the Escherichia coli yacK gene
|
|
70005
|
Oxidation of phenolate siderophores by the multicopper oxidase encoded by the Escherichia coli yacK gene
|
|
70006
|
Oxidation of phenolate siderophores by the multicopper oxidase encoded by the Escherichia coli yacK gene
|
|
70007
|
Oxidation of phenolate siderophores by the multicopper oxidase encoded by the Escherichia coli yacK gene
|
|
70008
|
Oxidation of phenolate siderophores by the multicopper oxidase encoded by the Escherichia coli yacK gene
|
|
70009
|
Oxidation of phenolate siderophores by the multicopper oxidase encoded by the Escherichia coli yacK gene
|
|
70010
|
Oxidation of phenolate siderophores by the multicopper oxidase encoded by the Escherichia coli yacK gene
|
|
70011
|
Oxidation of phenolate siderophores by the multicopper oxidase encoded by the Escherichia coli yacK gene
|
|
70012
|
Enhancement in catalytic activity of CotA-laccase from Bacillus pumilus W3 via site-directed mutagenesis
|
|
70013
|
Enhancement in catalytic activity of CotA-laccase from Bacillus pumilus W3 via site-directed mutagenesis
|
|
70014
|
Enhancement in catalytic activity of CotA-laccase from Bacillus pumilus W3 via site-directed mutagenesis
|
|
70015
|
Enhancement in catalytic activity of CotA-laccase from Bacillus pumilus W3 via site-directed mutagenesis
|
|
70016
|
Kinetic characterization of laccase from Bacillus atrophaeus, and its potential in juice clarification in free ...
|
|
70017
|
Kinetic characterization of laccase from Bacillus atrophaeus, and its potential in juice clarification in free ...
|
|
70018
|
Investigations of Accessibility of T2/T3 Copper Center of Two-Domain Laccase from Streptomyces ...
|
|
70019
|
Investigations of Accessibility of T2/T3 Copper Center of Two-Domain Laccase from Streptomyces ...
|
|
70020
|
Investigations of Accessibility of T2/T3 Copper Center of Two-Domain Laccase from Streptomyces ...
|
|
70021
|
Investigations of Accessibility of T2/T3 Copper Center of Two-Domain Laccase from Streptomyces ...
|
|
70022
|
Investigations of Accessibility of T2/T3 Copper Center of Two-Domain Laccase from Streptomyces ...
|
|
70023
|
Investigations of Accessibility of T2/T3 Copper Center of Two-Domain Laccase from Streptomyces ...
|
|
70024
|
Investigations of Accessibility of T2/T3 Copper Center of Two-Domain Laccase from Streptomyces ...
|
|
70025
|
Investigations of Accessibility of T2/T3 Copper Center of Two-Domain Laccase from Streptomyces ...
|
|
70026
|
CotA laccase, a novel aflatoxin oxidase from Bacillus licheniformis, transforms aflatoxin B1 to ...
|
|
70027
|
CotA laccase, a novel aflatoxin oxidase from Bacillus licheniformis, transforms aflatoxin B1 to ...
|
|
70028
|
CotA laccase, a novel aflatoxin oxidase from Bacillus licheniformis, transforms aflatoxin B1 to ...
|
|
70029
|
CotA laccase, a novel aflatoxin oxidase from Bacillus licheniformis, transforms aflatoxin B1 to ...
|
|
70030
|
Characterization of yellow bacterial laccase SmLac/role of redox mediators in azo dye decolorization
|
|
70031
|
Characterization of yellow bacterial laccase SmLac/role of redox mediators in azo dye decolorization
|
|
70032
|
Enhanced hydrolysis of lignocellulosic biomass with doping of a highly thermostable recombinant laccase
|
|
70033
|
Narrowing laccase substrate specificity using active site saturation mutagenesis
|
|
70034
|
Narrowing laccase substrate specificity using active site saturation mutagenesis
|
|
70035
|
Narrowing laccase substrate specificity using active site saturation mutagenesis
|
|
70036
|
Narrowing laccase substrate specificity using active site saturation mutagenesis
|
|
70037
|
Enzymatic biotransformation of the azo dye Sudan Orange G with bacterial CotA-laccase
|
|
70038
|
Enzymatic biotransformation of the azo dye Sudan Orange G with bacterial CotA-laccase
|
|
70039
|
Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural ...
|
|
70040
|
Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural ...
|
|
70041
|
Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural ...
|
|
70042
|
Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural ...
|
|
70043
|
Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural ...
|
|
70044
|
Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural ...
|
|
70045
|
Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural ...
|
|
70046
|
Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural ...
|
|
70047
|
Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural ...
|
|
70048
|
Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural ...
|
|
70049
|
Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural ...
|
|
70050
|
Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural ...
|
|
70051
|
A novel laccase from thermoalkaliphilic bacterium Caldalkalibacillus thermarum strain TA2.A1 able to catalyze ...
|
|
70052
|
Extracellular expression of mutant CotA-laccase SF in Escherichia coli and its degradation of malachite green
|
|
70053
|
Gene cloning, protein purification, and enzymatic properties of multicopper oxidase, from Klebsiella sp. 601
|
|
70054
|
Gene cloning, protein purification, and enzymatic properties of multicopper oxidase, from Klebsiella sp. 601
|
|
70055
|
Gene cloning, protein purification, and enzymatic properties of multicopper oxidase, from Klebsiella sp. 601
|
|
70056
|
A robust metallo-oxidase from the hyperthermophilic bacterium Aquifex aeolicus
|
|
70057
|
A robust metallo-oxidase from the hyperthermophilic bacterium Aquifex aeolicus
|
|
70058
|
A robust metallo-oxidase from the hyperthermophilic bacterium Aquifex aeolicus
|
|
70059
|
A robust metallo-oxidase from the hyperthermophilic bacterium Aquifex aeolicus
|
|
70060
|
A robust metallo-oxidase from the hyperthermophilic bacterium Aquifex aeolicus
|
|
70061
|
A robust metallo-oxidase from the hyperthermophilic bacterium Aquifex aeolicus
|
|
70062
|
A robust metallo-oxidase from the hyperthermophilic bacterium Aquifex aeolicus
|
|
70063
|
A robust metallo-oxidase from the hyperthermophilic bacterium Aquifex aeolicus
|
|
70064
|
A robust metallo-oxidase from the hyperthermophilic bacterium Aquifex aeolicus
|
|
70065
|
A robust metallo-oxidase from the hyperthermophilic bacterium Aquifex aeolicus
|
|
70066
|
A robust metallo-oxidase from the hyperthermophilic bacterium Aquifex aeolicus
|
|
70067
|
A robust metallo-oxidase from the hyperthermophilic bacterium Aquifex aeolicus
|
|
70068
|
A robust metallo-oxidase from the hyperthermophilic bacterium Aquifex aeolicus
|
|
70069
|
A robust metallo-oxidase from the hyperthermophilic bacterium Aquifex aeolicus
|
|
70070
|
A robust metallo-oxidase from the hyperthermophilic bacterium Aquifex aeolicus
|
|
70071
|
A robust metallo-oxidase from the hyperthermophilic bacterium Aquifex aeolicus
|
|
70072
|
Multiple Tolerance and Dye Decolorization Ability of a Novel Laccase Identified from Staphylococcus ...
|
|
70073
|
A hyperthermophilic laccase from Thermus thermophilus HB27
|
|
70074
|
A hyperthermophilic laccase from Thermus thermophilus HB27
|
|
70075
|
Insight into stability of CotA laccase from the spore coat of Bacillus subtilis
|
|
70076
|
Insight into stability of CotA laccase from the spore coat of Bacillus subtilis
|
|
70077
|
Insight into stability of CotA laccase from the spore coat of Bacillus subtilis
|
|
70078
|
Purification and kinetic characterization of human indoleamine 2,3-dioxygenases 1 and 2 (IDO1 and IDO2) and ...
|
|
70079
|
Purification and kinetic characterization of human indoleamine 2,3-dioxygenases 1 and 2 (IDO1 and IDO2) and ...
|
|
70080
|
Purification and kinetic characterization of human indoleamine 2,3-dioxygenases 1 and 2 (IDO1 and IDO2) and ...
|
|
70081
|
Purification and kinetic characterization of human indoleamine 2,3-dioxygenases 1 and 2 (IDO1 and IDO2) and ...
|
|
70082
|
Purification and kinetic characterization of human indoleamine 2,3-dioxygenases 1 and 2 (IDO1 and IDO2) and ...
|
|
70083
|
Purification and kinetic characterization of human indoleamine 2,3-dioxygenases 1 and 2 (IDO1 and IDO2) and ...
|
|
70084
|
Purification and kinetic characterization of human indoleamine 2,3-dioxygenases 1 and 2 (IDO1 and IDO2) and ...
|
|
70085
|
Site-directed mutation of a laccase from Thermus thermophilus: effect on the activity profile
|
|
70086
|
Site-directed mutation of a laccase from Thermus thermophilus: effect on the activity profile
|
|
70087
|
Site-directed mutation of a laccase from Thermus thermophilus: effect on the activity profile
|
|
70088
|
Site-directed mutation of a laccase from Thermus thermophilus: effect on the activity profile
|
|
70089
|
Desulfovibrio DA2_CueO is a novel multicopper oxidase with cuprous, ferrous and phenol oxidase activity
|
|
70090
|
Desulfovibrio DA2_CueO is a novel multicopper oxidase with cuprous, ferrous and phenol oxidase activity
|
|
70091
|
Desulfovibrio DA2_CueO is a novel multicopper oxidase with cuprous, ferrous and phenol oxidase activity
|
|
70092
|
Desulfovibrio DA2_CueO is a novel multicopper oxidase with cuprous, ferrous and phenol oxidase activity
|
|
70093
|
Biochemical Characteristics of Three Laccase Isoforms from the Basidiomycete Pleurotus nebrodensis
|
|
70094
|
Biochemical Characteristics of Three Laccase Isoforms from the Basidiomycete Pleurotus nebrodensis
|
|
70095
|
Biochemical Characteristics of Three Laccase Isoforms from the Basidiomycete Pleurotus nebrodensis
|
|
70096
|
Purification and Characterization of a White Laccase with Pronounced Dye Decolorizing Ability and HIV-1 ...
|
|
70097
|
Purification and Characterization of a White Laccase with Pronounced Dye Decolorizing Ability and HIV-1 ...
|
|
70098
|
Purification and biochemical characterization of a new alkali-stable laccase from Trametes sp. isolated in ...
|
|
70099
|
Purification and biochemical characterization of a new alkali-stable laccase from Trametes sp. isolated in ...
|
|
70100
|
Enzyme-catalyzed acylation of homoserine: mechanistic characterization of the Escherichia coli metA-encoded ...
|
|
70101
|
Enzyme-catalyzed acylation of homoserine: mechanistic characterization of the Escherichia coli metA-encoded ...
|
|
70102
|
Enzyme-catalyzed acylation of homoserine: mechanistic characterization of the Escherichia coli metA-encoded ...
|
|
70103
|
Enzyme-catalyzed acylation of homoserine: mechanistic characterization of the Escherichia coli metA-encoded ...
|
|
70104
|
Enzyme-catalyzed acylation of homoserine: mechanistic characterization of the Escherichia coli metA-encoded ...
|
|
70105
|
Enzyme-catalyzed acylation of homoserine: mechanistic characterization of the Escherichia coli metA-encoded ...
|
|
70106
|
Enzyme-catalyzed acylation of homoserine: mechanistic characterization of the Escherichia coli metA-encoded ...
|
|
70107
|
The biochemical consequences of alpha2,6(N) sialyltransferase induction by dexamethasone on sialoglycoprotein ...
|
|
70108
|
The biochemical consequences of alpha2,6(N) sialyltransferase induction by dexamethasone on sialoglycoprotein ...
|
|
70109
|
The biochemical consequences of alpha2,6(N) sialyltransferase induction by dexamethasone on sialoglycoprotein ...
|
|
70110
|
The biochemical consequences of alpha2,6(N) sialyltransferase induction by dexamethasone on sialoglycoprotein ...
|
|
70111
|
Hydrogen Peroxide-Resistant CotA and YjqC of Bacillus altitudinis Spores Are a Promising Biocatalyst for ...
|
|
70112
|
Hydrogen Peroxide-Resistant CotA and YjqC of Bacillus altitudinis Spores Are a Promising Biocatalyst for ...
|
|
70113
|
Hydrogen Peroxide-Resistant CotA and YjqC of Bacillus altitudinis Spores Are a Promising Biocatalyst for ...
|
|
70114
|
Hydrogen Peroxide-Resistant CotA and YjqC of Bacillus altitudinis Spores Are a Promising Biocatalyst for ...
|
|
70115
|
Hydrogen Peroxide-Resistant CotA and YjqC of Bacillus altitudinis Spores Are a Promising Biocatalyst for ...
|
|
70116
|
Hydrogen Peroxide-Resistant CotA and YjqC of Bacillus altitudinis Spores Are a Promising Biocatalyst for ...
|
|
70117
|
Hydrogen Peroxide-Resistant CotA and YjqC of Bacillus altitudinis Spores Are a Promising Biocatalyst for ...
|
|
70118
|
Hydrogen Peroxide-Resistant CotA and YjqC of Bacillus altitudinis Spores Are a Promising Biocatalyst for ...
|
|
70119
|
Hydrogen Peroxide-Resistant CotA and YjqC of Bacillus altitudinis Spores Are a Promising Biocatalyst for ...
|
|
70120
|
Hydrogen Peroxide-Resistant CotA and YjqC of Bacillus altitudinis Spores Are a Promising Biocatalyst for ...
|
|
70121
|
Hydrogen Peroxide-Resistant CotA and YjqC of Bacillus altitudinis Spores Are a Promising Biocatalyst for ...
|
|
70122
|
Hydrogen Peroxide-Resistant CotA and YjqC of Bacillus altitudinis Spores Are a Promising Biocatalyst for ...
|
|
70123
|
Hydrogen Peroxide-Resistant CotA and YjqC of Bacillus altitudinis Spores Are a Promising Biocatalyst for ...
|
|
70124
|
Hydrogen Peroxide-Resistant CotA and YjqC of Bacillus altitudinis Spores Are a Promising Biocatalyst for ...
|
|
70125
|
Hydrogen Peroxide-Resistant CotA and YjqC of Bacillus altitudinis Spores Are a Promising Biocatalyst for ...
|
|
70126
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70127
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70128
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70129
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70130
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70131
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70132
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70133
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70134
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70135
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70136
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70137
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70138
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70139
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70140
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70141
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70142
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70143
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70144
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70145
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70146
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70147
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70148
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70149
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70150
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70151
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70152
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70153
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70154
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70155
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70156
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70157
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70158
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70159
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70160
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70161
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70162
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70163
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70164
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70165
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70166
|
Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical ...
|
|
70167
|
Transformation of low molecular compounds and soil humic acid by two domain laccase of Streptomyces puniceus ...
|
|
70168
|
Transformation of low molecular compounds and soil humic acid by two domain laccase of Streptomyces puniceus ...
|
|
70169
|
Transformation of low molecular compounds and soil humic acid by two domain laccase of Streptomyces puniceus ...
|
|
70170
|
Transformation of low molecular compounds and soil humic acid by two domain laccase of Streptomyces puniceus ...
|
|
70171
|
Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the ...
|
|
70172
|
Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the ...
|
|
70173
|
Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the ...
|
|
70174
|
Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the ...
|
|
70175
|
Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the ...
|
|
70176
|
Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the ...
|
|
70177
|
Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the ...
|
|
70178
|
Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the ...
|
|
70179
|
Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the ...
|
|
70180
|
Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the ...
|
|
70181
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70182
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70183
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70184
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70185
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70186
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70187
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70188
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70189
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70190
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70191
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70192
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70193
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70194
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70195
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70196
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70197
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70198
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70199
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70200
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70201
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70202
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70203
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70204
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70205
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70206
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70207
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70208
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70209
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70210
|
Comparison of different microbial laccases as tools for industrial uses
|
|
70211
|
A novel laccase with urate oxidation activity from Lysobacter sp. T-15.
|
|
70212
|
A novel laccase with urate oxidation activity from Lysobacter sp. T-15.
|
|
70213
|
A novel laccase with urate oxidation activity from Lysobacter sp. T-15.
|
|
70214
|
A novel laccase with urate oxidation activity from Lysobacter sp. T-15.
|
|
70215
|
A novel laccase with urate oxidation activity from Lysobacter sp. T-15.
|
|
70216
|
A novel laccase with urate oxidation activity from Lysobacter sp. T-15.
|
|
70217
|
A novel laccase with urate oxidation activity from Lysobacter sp. T-15.
|
|
70218
|
A novel laccase with urate oxidation activity from Lysobacter sp. T-15.
|
|
70219
|
A novel laccase with urate oxidation activity from Lysobacter sp. T-15.
|
|
70220
|
The multi-copper-ion oxidase CueO of Salmonella enterica serovar Typhimurium is required for systemic ...
|
|
70221
|
The multi-copper-ion oxidase CueO of Salmonella enterica serovar Typhimurium is required for systemic ...
|
|
70222
|
Site-Directed Mutagenesis of Multicopper Oxidase from Hyperthermophilic Archaea for High-Voltage Biofuel Cells
|
|
70223
|
Site-Directed Mutagenesis of Multicopper Oxidase from Hyperthermophilic Archaea for High-Voltage Biofuel Cells
|
|
70224
|
Site-Directed Mutagenesis of Multicopper Oxidase from Hyperthermophilic Archaea for High-Voltage Biofuel Cells
|
|
70225
|
Enhancing the decolorization activity of Bacillus pumilus W3 CotA-laccase to Reactive Black 5 by site- ...
|
|
70226
|
Enhancing the decolorization activity of Bacillus pumilus W3 CotA-laccase to Reactive Black 5 by site- ...
|
|
70227
|
Enhancing the decolorization activity of Bacillus pumilus W3 CotA-laccase to Reactive Black 5 by site- ...
|
|
70228
|
Enhancing the decolorization activity of Bacillus pumilus W3 CotA-laccase to Reactive Black 5 by site- ...
|
|
70229
|
Enhancing the decolorization activity of Bacillus pumilus W3 CotA-laccase to Reactive Black 5 by site- ...
|
|
70230
|
Enhancing the decolorization activity of Bacillus pumilus W3 CotA-laccase to Reactive Black 5 by site- ...
|
|
70231
|
Enhancing the decolorization activity of Bacillus pumilus W3 CotA-laccase to Reactive Black 5 by site- ...
|
|
70232
|
Enhancing the decolorization activity of Bacillus pumilus W3 CotA-laccase to Reactive Black 5 by site- ...
|
|
70233
|
Enhancing the decolorization activity of Bacillus pumilus W3 CotA-laccase to Reactive Black 5 by site- ...
|
|
70234
|
Enhancing the decolorization activity of Bacillus pumilus W3 CotA-laccase to Reactive Black 5 by site- ...
|
|
70235
|
Enhancing the decolorization activity of Bacillus pumilus W3 CotA-laccase to Reactive Black 5 by site- ...
|
|
70236
|
Enhancing the decolorization activity of Bacillus pumilus W3 CotA-laccase to Reactive Black 5 by site- ...
|
|
70237
|
Novel polyphenol oxidase mined from a metagenome expression library of bovine rumen: biochemical properties, ...
|
|
70238
|
Novel polyphenol oxidase mined from a metagenome expression library of bovine rumen: biochemical properties, ...
|
|
70239
|
Novel polyphenol oxidase mined from a metagenome expression library of bovine rumen: biochemical properties, ...
|
|
70240
|
Novel polyphenol oxidase mined from a metagenome expression library of bovine rumen: biochemical properties, ...
|
|
70241
|
Novel polyphenol oxidase mined from a metagenome expression library of bovine rumen: biochemical properties, ...
|
|
70242
|
Novel polyphenol oxidase mined from a metagenome expression library of bovine rumen: biochemical properties, ...
|
|
70243
|
Novel polyphenol oxidase mined from a metagenome expression library of bovine rumen: biochemical properties, ...
|
|
70244
|
The second conserved motif in bacterial laccase regulates catalysis and robustness
|
|
70245
|
The second conserved motif in bacterial laccase regulates catalysis and robustness
|
|
70246
|
The second conserved motif in bacterial laccase regulates catalysis and robustness
|
|
70247
|
The second conserved motif in bacterial laccase regulates catalysis and robustness
|
|
70248
|
The second conserved motif in bacterial laccase regulates catalysis and robustness
|
|
70249
|
The second conserved motif in bacterial laccase regulates catalysis and robustness
|
|
70250
|
The second conserved motif in bacterial laccase regulates catalysis and robustness
|
|
70251
|
The second conserved motif in bacterial laccase regulates catalysis and robustness
|
|
70252
|
The second conserved motif in bacterial laccase regulates catalysis and robustness
|
|
70253
|
The second conserved motif in bacterial laccase regulates catalysis and robustness
|
|
70254
|
The second conserved motif in bacterial laccase regulates catalysis and robustness
|
|
70255
|
The second conserved motif in bacterial laccase regulates catalysis and robustness
|
|
70256
|
The second conserved motif in bacterial laccase regulates catalysis and robustness
|
|
70257
|
The second conserved motif in bacterial laccase regulates catalysis and robustness
|
|
70258
|
The second conserved motif in bacterial laccase regulates catalysis and robustness
|
|
70259
|
The second conserved motif in bacterial laccase regulates catalysis and robustness
|
|
70260
|
Sialyltransferase activity of human plasma and aortic intima is enhanced in atherosclerosis
|
|
70261
|
Sialyltransferase activity of human plasma and aortic intima is enhanced in atherosclerosis
|
|
70262
|
Sialyltransferase activity of human plasma and aortic intima is enhanced in atherosclerosis
|
|
70263
|
Sialyltransferase activity of human plasma and aortic intima is enhanced in atherosclerosis
|
|
70264
|
Purification and properties of rabbit spermatozoal acrosomal neuraminidase.
|
|
70265
|
Purification and properties of rabbit spermatozoal acrosomal neuraminidase.
|
|
70266
|
Purification and properties of rabbit spermatozoal acrosomal neuraminidase.
|
|
70267
|
Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis
|
|
70268
|
Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis
|
|
70269
|
Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis
|
|
70270
|
Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis
|
|
70271
|
Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis
|
|
70272
|
Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis
|
|
70273
|
Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis
|
|
70274
|
Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis
|
|
70275
|
Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis
|
|
70276
|
Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis
|
|
70277
|
Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis
|
|
70278
|
Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis
|
|
70279
|
Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis
|
|
70280
|
Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis
|
|
70281
|
Analysis of a chemical plant defense mechanism in grasses.
|
|
70282
|
Tryptophan pyrrolase of rabbit intestine. D- and L-tryptophan-cleaving enzyme or enzymes
|
|
70283
|
Tryptophan pyrrolase of rabbit intestine. D- and L-tryptophan-cleaving enzyme or enzymes
|
|
70284
|
Tryptophan pyrrolase of rabbit intestine. D- and L-tryptophan-cleaving enzyme or enzymes
|
|
70285
|
Tryptophan pyrrolase of rabbit intestine. D- and L-tryptophan-cleaving enzyme or enzymes
|
|
70286
|
Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor ...
|
|
70287
|
Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor ...
|
|
70288
|
Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor ...
|
|
70289
|
Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor ...
|
|
70290
|
Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor ...
|
|
70291
|
Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor ...
|
|
70292
|
Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor ...
|
|
70293
|
Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor ...
|
|
70294
|
Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor ...
|
|
70295
|
Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor ...
|
|
70296
|
Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor ...
|
|
70297
|
Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor ...
|
|
70298
|
Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor ...
|
|
70299
|
Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor ...
|
|
70300
|
Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor ...
|
|
70301
|
Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor ...
|
|
70302
|
Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor ...
|
|
70303
|
Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor ...
|
|
70304
|
Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor ...
|
|
70305
|
Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor ...
|
|
70306
|
Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor ...
|
|
70307
|
Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor ...
|
|
70308
|
Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor ...
|
|
70309
|
Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor ...
|
|
70310
|
Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor ...
|
|
70311
|
Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor ...
|
|
70312
|
Regulation of plant cytosolic aldolase functions by redox-modifications
|
|
70313
|
Regulation of plant cytosolic aldolase functions by redox-modifications
|





 If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.
If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.