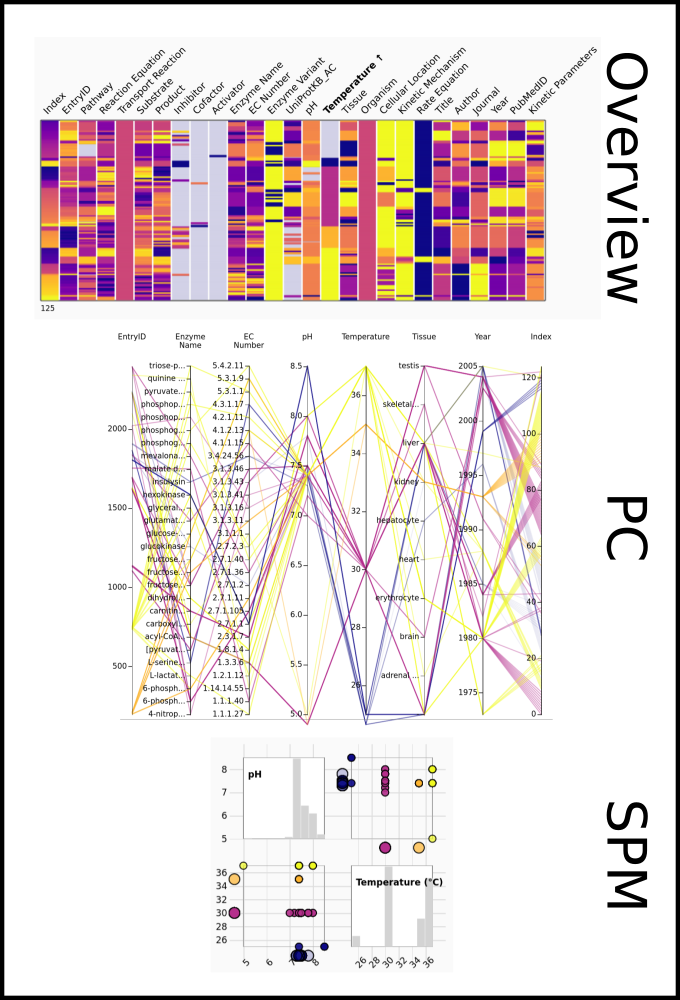
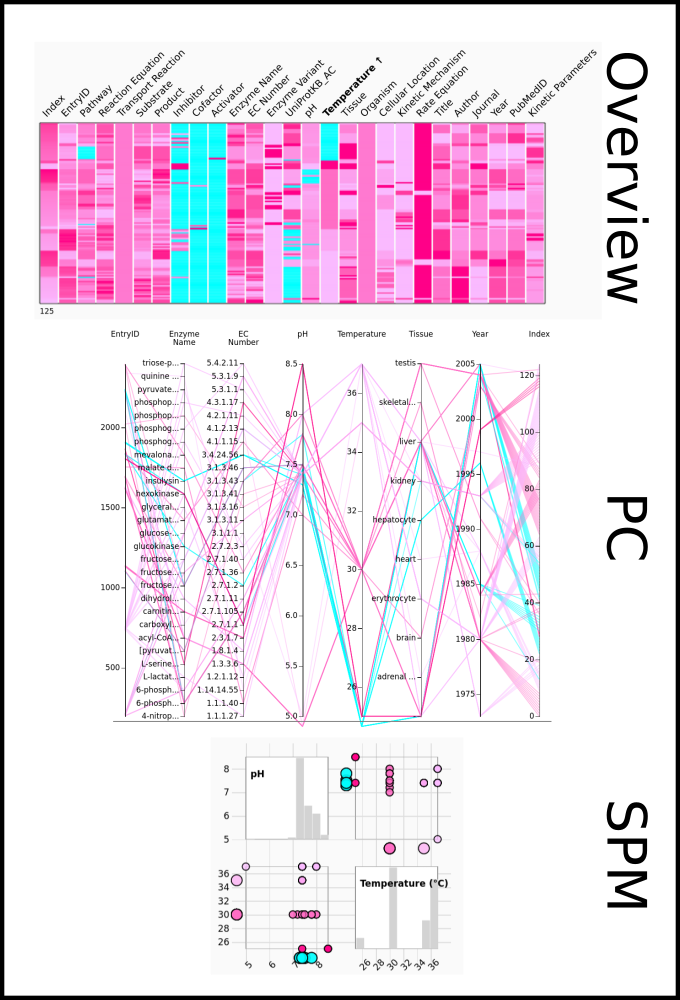
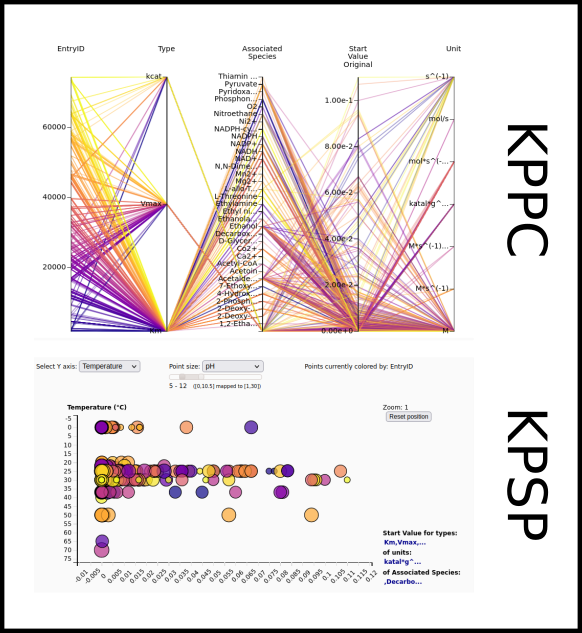
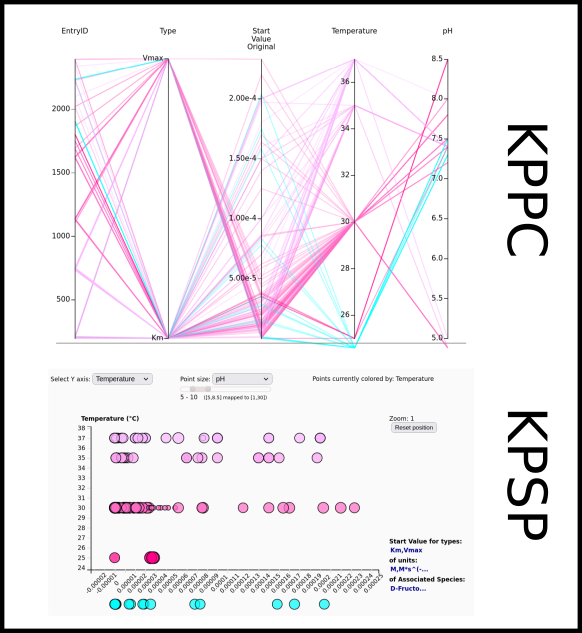
To improve SABIO-RK data interoperability, semantic markup was added to web pages as described and defined by
This structured information makes it easier to discover, collate and analyse our data.
|
69001
|
Catalysis, stereochemistry, and inhibition of ureidoglycolate lyase
|
|
69002
|
Catalysis, stereochemistry, and inhibition of ureidoglycolate lyase
|
|
69003
|
Catalysis, stereochemistry, and inhibition of ureidoglycolate lyase
|
|
69004
|
Catalysis, stereochemistry, and inhibition of ureidoglycolate lyase
|
|
69005
|
Catalysis, stereochemistry, and inhibition of ureidoglycolate lyase
|
|
69006
|
Catalysis, stereochemistry, and inhibition of ureidoglycolate lyase
|
|
69007
|
Catalysis, stereochemistry, and inhibition of ureidoglycolate lyase
|
|
69008
|
Catalysis, stereochemistry, and inhibition of ureidoglycolate lyase
|
|
69009
|
Catalysis, stereochemistry, and inhibition of ureidoglycolate lyase
|
|
69010
|
Catalysis, stereochemistry, and inhibition of ureidoglycolate lyase
|
|
69011
|
Catalysis, stereochemistry, and inhibition of ureidoglycolate lyase
|
|
69012
|
Catalysis, stereochemistry, and inhibition of ureidoglycolate lyase
|
|
69013
|
Catalysis, stereochemistry, and inhibition of ureidoglycolate lyase
|
|
69014
|
Catalysis, stereochemistry, and inhibition of ureidoglycolate lyase
|
|
69015
|
Catalysis, stereochemistry, and inhibition of ureidoglycolate lyase
|
|
69016
|
Catalysis, stereochemistry, and inhibition of ureidoglycolate lyase
|
|
69017
|
Catalysis, stereochemistry, and inhibition of ureidoglycolate lyase
|
|
69018
|
Catalysis, stereochemistry, and inhibition of ureidoglycolate lyase
|
|
69019
|
Catalysis, stereochemistry, and inhibition of ureidoglycolate lyase
|
|
69020
|
Functional differences between rabbit and human Na(+)-dicarboxylate cotransporters, NaDC-1 and hNaDC-1.
|
|
69021
|
Functional differences between rabbit and human Na(+)-dicarboxylate cotransporters, NaDC-1 and hNaDC-1.
|
|
69022
|
Functional differences between rabbit and human Na(+)-dicarboxylate cotransporters, NaDC-1 and hNaDC-1.
|
|
69023
|
Functional differences between rabbit and human Na(+)-dicarboxylate cotransporters, NaDC-1 and hNaDC-1.
|
|
69024
|
Functional differences between rabbit and human Na(+)-dicarboxylate cotransporters, NaDC-1 and hNaDC-1.
|
|
69025
|
Functional differences between rabbit and human Na(+)-dicarboxylate cotransporters, NaDC-1 and hNaDC-1.
|
|
69026
|
Functional differences between rabbit and human Na(+)-dicarboxylate cotransporters, NaDC-1 and hNaDC-1.
|
|
69027
|
Functional differences between rabbit and human Na(+)-dicarboxylate cotransporters, NaDC-1 and hNaDC-1.
|
|
69028
|
Functional differences between rabbit and human Na(+)-dicarboxylate cotransporters, NaDC-1 and hNaDC-1.
|
|
69029
|
Functional differences between rabbit and human Na(+)-dicarboxylate cotransporters, NaDC-1 and hNaDC-1.
|
|
69030
|
Functional differences between rabbit and human Na(+)-dicarboxylate cotransporters, NaDC-1 and hNaDC-1.
|
|
69031
|
Functional differences between rabbit and human Na(+)-dicarboxylate cotransporters, NaDC-1 and hNaDC-1.
|
|
69032
|
Functional differences between rabbit and human Na(+)-dicarboxylate cotransporters, NaDC-1 and hNaDC-1.
|
|
69033
|
Functional differences between rabbit and human Na(+)-dicarboxylate cotransporters, NaDC-1 and hNaDC-1.
|
|
69034
|
Functional differences between rabbit and human Na(+)-dicarboxylate cotransporters, NaDC-1 and hNaDC-1.
|
|
69035
|
Isoform-dependent differences in feedback regulation and subcellular localization of serine acetyltransferase ...
|
|
69036
|
Isoform-dependent differences in feedback regulation and subcellular localization of serine acetyltransferase ...
|
|
69037
|
Isoform-dependent differences in feedback regulation and subcellular localization of serine acetyltransferase ...
|
|
69038
|
Isoform-dependent differences in feedback regulation and subcellular localization of serine acetyltransferase ...
|
|
69039
|
Isoform-dependent differences in feedback regulation and subcellular localization of serine acetyltransferase ...
|
|
69040
|
Isoform-dependent differences in feedback regulation and subcellular localization of serine acetyltransferase ...
|
|
69041
|
New findings on SNP variants of human protein L-isoaspartyl methyltransferase that affect catalytic activity, ...
|
|
69042
|
New findings on SNP variants of human protein L-isoaspartyl methyltransferase that affect catalytic activity, ...
|
|
69043
|
New findings on SNP variants of human protein L-isoaspartyl methyltransferase that affect catalytic activity, ...
|
|
69044
|
New findings on SNP variants of human protein L-isoaspartyl methyltransferase that affect catalytic activity, ...
|
|
69045
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69046
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69047
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69048
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69049
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69050
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69051
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69052
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69053
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69054
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69055
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69056
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69057
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69058
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69059
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69060
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69061
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69062
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69063
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69064
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69065
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69066
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69067
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69068
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69069
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69070
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69071
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69072
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69073
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69074
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69075
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69076
|
Roles of amino acid residues surrounding phosphorylation site 1 of branched-chain alpha-ketoacid dehydrogenase ...
|
|
69077
|
Differential substrate behaviour of phenol and aniline derivatives during oxidation by horseradish peroxidase: ...
|
|
69078
|
Differential substrate behaviour of phenol and aniline derivatives during oxidation by horseradish peroxidase: ...
|
|
69079
|
Differential substrate behaviour of phenol and aniline derivatives during oxidation by horseradish peroxidase: ...
|
|
69080
|
Differential substrate behaviour of phenol and aniline derivatives during oxidation by horseradish peroxidase: ...
|
|
69081
|
Differential substrate behaviour of phenol and aniline derivatives during oxidation by horseradish peroxidase: ...
|
|
69082
|
Differential substrate behaviour of phenol and aniline derivatives during oxidation by horseradish peroxidase: ...
|
|
69083
|
Differential substrate behaviour of phenol and aniline derivatives during oxidation by horseradish peroxidase: ...
|
|
69084
|
Differential substrate behaviour of phenol and aniline derivatives during oxidation by horseradish peroxidase: ...
|
|
69085
|
Differential substrate behaviour of phenol and aniline derivatives during oxidation by horseradish peroxidase: ...
|
|
69086
|
Differential substrate behaviour of phenol and aniline derivatives during oxidation by horseradish peroxidase: ...
|
|
69087
|
Differential substrate behaviour of phenol and aniline derivatives during oxidation by horseradish peroxidase: ...
|
|
69088
|
Differential substrate behaviour of phenol and aniline derivatives during oxidation by horseradish peroxidase: ...
|
|
69089
|
Differential substrate behaviour of phenol and aniline derivatives during oxidation by horseradish peroxidase: ...
|
|
69090
|
Differential substrate behaviour of phenol and aniline derivatives during oxidation by horseradish peroxidase: ...
|
|
69091
|
Purification of bacterial L-methionine gamma-lyase
|
|
69092
|
Trifluoromethionine, a prodrug designed against methionine gamma-lyase-containing pathogens, has efficacy in ...
|
|
69093
|
Trifluoromethionine, a prodrug designed against methionine gamma-lyase-containing pathogens, has efficacy in ...
|
|
69094
|
Isolation and purification of L-methionine-alpha-deamino-gamma-mercaptomethane-lyase (L-methioninase) from ...
|
|
69095
|
cDNA cloning, overexpression in Escherichia coli, purification and characterization of sheep liver cytosolic ...
|
|
69096
|
Purification and characterization of recombinant rabbit cytosolic serine hydroxymethyltransferase
|
|
69097
|
Purification and characterization of recombinant rabbit cytosolic serine hydroxymethyltransferase
|
|
69098
|
Purification and characterization of recombinant rabbit cytosolic serine hydroxymethyltransferase
|
|
69099
|
The first committed step reaction of caffeine biosynthesis: 7-methylxanthosine synthase is closely homologous ...
|
|
69100
|
Overexpression and characterization of dimeric and tetrameric forms of recombinant serine ...
|
|
69101
|
Overexpression and characterization of dimeric and tetrameric forms of recombinant serine ...
|
|
69102
|
Overexpression and characterization of dimeric and tetrameric forms of recombinant serine ...
|
|
69103
|
Overexpression and characterization of dimeric and tetrameric forms of recombinant serine ...
|
|
69104
|
Overexpression and characterization of dimeric and tetrameric forms of recombinant serine ...
|
|
69105
|
Overexpression and characterization of dimeric and tetrameric forms of recombinant serine ...
|
|
69106
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69107
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69108
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69109
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69110
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69111
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69112
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69113
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69114
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69115
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69116
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69117
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69118
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69119
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69120
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69121
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69122
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69123
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69124
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69125
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69126
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69127
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69128
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69129
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69130
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69131
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69132
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69133
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69134
|
Identification of Mn2+-binding aspartates from alpha, beta, and gamma subunits of human NAD-dependent ...
|
|
69135
|
A comparison of the soluble and mitochondrial forms of human isocitrate dehydrogenase and an examination of ...
|
|
69136
|
A comparison of the soluble and mitochondrial forms of human isocitrate dehydrogenase and an examination of ...
|
|
69137
|
A comparison of the soluble and mitochondrial forms of human isocitrate dehydrogenase and an examination of ...
|
|
69138
|
A comparison of the soluble and mitochondrial forms of human isocitrate dehydrogenase and an examination of ...
|
|
69139
|
Characterization of Plasmodium falciparum serine hydroxymethyltransferase-A potential antimalarial target
|
|
69140
|
Characterization of Plasmodium falciparum serine hydroxymethyltransferase-A potential antimalarial target
|
|
69141
|
Characterization of Plasmodium falciparum serine hydroxymethyltransferase-A potential antimalarial target
|
|
69142
|
Characterization of Plasmodium falciparum serine hydroxymethyltransferase-A potential antimalarial target
|
|
69143
|
Characterization of Plasmodium falciparum serine hydroxymethyltransferase-A potential antimalarial target
|
|
69144
|
Characterization of Plasmodium falciparum serine hydroxymethyltransferase-A potential antimalarial target
|
|
69145
|
Characterization of Plasmodium falciparum serine hydroxymethyltransferase-A potential antimalarial target
|
|
69146
|
Characterization of Plasmodium falciparum serine hydroxymethyltransferase-A potential antimalarial target
|
|
69147
|
Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate ...
|
|
69148
|
Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate ...
|
|
69149
|
Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate ...
|
|
69150
|
Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate ...
|
|
69151
|
Ca(2+) modulates respiratory and steroidogenic activities of human term placental mitochondria.
|
|
69152
|
Ca(2+) modulates respiratory and steroidogenic activities of human term placental mitochondria.
|
|
69153
|
Ca(2+) modulates respiratory and steroidogenic activities of human term placental mitochondria.
|
|
69154
|
Ca(2+) modulates respiratory and steroidogenic activities of human term placental mitochondria.
|
|
69155
|
Ca(2+) modulates respiratory and steroidogenic activities of human term placental mitochondria.
|
|
69156
|
Ca(2+) modulates respiratory and steroidogenic activities of human term placental mitochondria.
|
|
69157
|
Ca(2+) modulates respiratory and steroidogenic activities of human term placental mitochondria.
|
|
69158
|
Ca(2+) modulates respiratory and steroidogenic activities of human term placental mitochondria.
|
|
69159
|
Ca(2+) modulates respiratory and steroidogenic activities of human term placental mitochondria.
|
|
69160
|
Ca(2+) modulates respiratory and steroidogenic activities of human term placental mitochondria.
|
|
69161
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69162
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69163
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69164
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69165
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69166
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69167
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69168
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69169
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69170
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69171
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69172
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69173
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69174
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69175
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69176
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69177
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69178
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69179
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69180
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69181
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69182
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69183
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69184
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69185
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69186
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69187
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69188
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69189
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69190
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69191
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69192
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69193
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69194
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69195
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69196
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69197
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69198
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69199
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69200
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69201
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69202
|
A comparative study of the regulation of Ca2+ of the activities of the 2-oxoglutarate dehydrogenase complex ...
|
|
69203
|
The alpha-glucuronidase, GlcA67A, of Cellvibrio japonicus utilizes the carboxylate and methyl groups of ...
|
|
69204
|
The alpha-glucuronidase, GlcA67A, of Cellvibrio japonicus utilizes the carboxylate and methyl groups of ...
|
|
69205
|
The alpha-glucuronidase, GlcA67A, of Cellvibrio japonicus utilizes the carboxylate and methyl groups of ...
|
|
69206
|
The alpha-glucuronidase, GlcA67A, of Cellvibrio japonicus utilizes the carboxylate and methyl groups of ...
|
|
69207
|
The alpha-glucuronidase, GlcA67A, of Cellvibrio japonicus utilizes the carboxylate and methyl groups of ...
|
|
69208
|
The alpha-glucuronidase, GlcA67A, of Cellvibrio japonicus utilizes the carboxylate and methyl groups of ...
|
|
69209
|
The alpha-glucuronidase, GlcA67A, of Cellvibrio japonicus utilizes the carboxylate and methyl groups of ...
|
|
69210
|
The alpha-glucuronidase, GlcA67A, of Cellvibrio japonicus utilizes the carboxylate and methyl groups of ...
|
|
69211
|
The alpha-glucuronidase, GlcA67A, of Cellvibrio japonicus utilizes the carboxylate and methyl groups of ...
|
|
69212
|
The alpha-glucuronidase, GlcA67A, of Cellvibrio japonicus utilizes the carboxylate and methyl groups of ...
|
|
69213
|
The alpha-glucuronidase, GlcA67A, of Cellvibrio japonicus utilizes the carboxylate and methyl groups of ...
|
|
69214
|
The alpha-glucuronidase, GlcA67A, of Cellvibrio japonicus utilizes the carboxylate and methyl groups of ...
|
|
69215
|
The alpha-glucuronidase, GlcA67A, of Cellvibrio japonicus utilizes the carboxylate and methyl groups of ...
|
|
69216
|
The alpha-glucuronidase, GlcA67A, of Cellvibrio japonicus utilizes the carboxylate and methyl groups of ...
|
|
69217
|
The alpha-glucuronidase, GlcA67A, of Cellvibrio japonicus utilizes the carboxylate and methyl groups of ...
|
|
69218
|
The alpha-glucuronidase, GlcA67A, of Cellvibrio japonicus utilizes the carboxylate and methyl groups of ...
|
|
69219
|
The alpha-glucuronidase, GlcA67A, of Cellvibrio japonicus utilizes the carboxylate and methyl groups of ...
|
|
69220
|
The plant biotin synthase reaction. Identification and characterization of essential mitochondrial accessory ...
|
|
69221
|
The plant biotin synthase reaction. Identification and characterization of essential mitochondrial accessory ...
|
|
69222
|
Characterization of a novel high-pH-tolerant laccase-like multicopper oxidase and its sequence diversity in ...
|
|
69223
|
Characterization of a novel high-pH-tolerant laccase-like multicopper oxidase and its sequence diversity in ...
|
|
69224
|
Different recombinant forms of polyphenol oxidase A, a laccase from Marinomonas mediterranea
|
|
69225
|
Different recombinant forms of polyphenol oxidase A, a laccase from Marinomonas mediterranea
|
|
69226
|
Different recombinant forms of polyphenol oxidase A, a laccase from Marinomonas mediterranea
|
|
69227
|
Different recombinant forms of polyphenol oxidase A, a laccase from Marinomonas mediterranea
|
|
69228
|
Different recombinant forms of polyphenol oxidase A, a laccase from Marinomonas mediterranea
|
|
69229
|
Different recombinant forms of polyphenol oxidase A, a laccase from Marinomonas mediterranea
|
|
69230
|
Different recombinant forms of polyphenol oxidase A, a laccase from Marinomonas mediterranea
|
|
69231
|
Different recombinant forms of polyphenol oxidase A, a laccase from Marinomonas mediterranea
|
|
69232
|
Different recombinant forms of polyphenol oxidase A, a laccase from Marinomonas mediterranea
|
|
69233
|
Different recombinant forms of polyphenol oxidase A, a laccase from Marinomonas mediterranea
|
|
69234
|
Different recombinant forms of polyphenol oxidase A, a laccase from Marinomonas mediterranea
|
|
69235
|
Different recombinant forms of polyphenol oxidase A, a laccase from Marinomonas mediterranea
|
|
69236
|
Increasing the catalytic activity of Bilirubin oxidase from Bacillus pumilus: Importance of host strain and ...
|
|
69237
|
Increasing the catalytic activity of Bilirubin oxidase from Bacillus pumilus: Importance of host strain and ...
|
|
69238
|
Purification and characterization of laccase from Sinorhizobium meliloti and analysis of the lacc gene
|
|
69239
|
Purification and characterization of laccase from Sinorhizobium meliloti and analysis of the lacc gene
|
|
69240
|
Purification and characterization of laccase from Sinorhizobium meliloti and analysis of the lacc gene
|
|
69241
|
Purification and characterization of laccase from Sinorhizobium meliloti and analysis of the lacc gene
|
|
69242
|
Purification and characterization of a recombinant laccase-like multi-copper oxidase from Paenibacillus ...
|
|
69243
|
Purification and characterization of a recombinant laccase-like multi-copper oxidase from Paenibacillus ...
|
|
69244
|
Purification and characterization of a recombinant laccase-like multi-copper oxidase from Paenibacillus ...
|
|
69245
|
Structural insight into the oxidation of sinapic acid by CotA laccase
|
|
69246
|
Structural insight into the oxidation of sinapic acid by CotA laccase
|
|
69247
|
Structural insight into the oxidation of sinapic acid by CotA laccase
|
|
69248
|
Structural insight into the oxidation of sinapic acid by CotA laccase
|
|
69249
|
Structural insight into the oxidation of sinapic acid by CotA laccase
|
|
69250
|
Structural insight into the oxidation of sinapic acid by CotA laccase
|
|
69251
|
Structural insight into the oxidation of sinapic acid by CotA laccase
|
|
69252
|
Structural insight into the oxidation of sinapic acid by CotA laccase
|
|
69253
|
Structural insight into the oxidation of sinapic acid by CotA laccase
|
|
69254
|
Purification and characterization of ornithine acetyltransferase from Saccharomyces cerevisiae.
|
|
69255
|
Purification and characterization of ornithine acetyltransferase from Saccharomyces cerevisiae.
|
|
69256
|
Solution structure of human peptidyl prolyl isomerase-like protein 1 and insights into its interaction with ...
|
|
69257
|
Solution structure of human peptidyl prolyl isomerase-like protein 1 and insights into its interaction with ...
|
|
69258
|
Homologs of the Rml enzymes from Salmonella enterica are responsible for dTDP-beta-L-rhamnose biosynthesis in ...
|
|
69259
|
Homologs of the Rml enzymes from Salmonella enterica are responsible for dTDP-beta-L-rhamnose biosynthesis in ...
|
|
69260
|
Homologs of the Rml enzymes from Salmonella enterica are responsible for dTDP-beta-L-rhamnose biosynthesis in ...
|
|
69261
|
Homologs of the Rml enzymes from Salmonella enterica are responsible for dTDP-beta-L-rhamnose biosynthesis in ...
|
|
69262
|
Homologs of the Rml enzymes from Salmonella enterica are responsible for dTDP-beta-L-rhamnose biosynthesis in ...
|
|
69263
|
Homologs of the Rml enzymes from Salmonella enterica are responsible for dTDP-beta-L-rhamnose biosynthesis in ...
|
|
69264
|
Homologs of the Rml enzymes from Salmonella enterica are responsible for dTDP-beta-L-rhamnose biosynthesis in ...
|
|
69265
|
Homologs of the Rml enzymes from Salmonella enterica are responsible for dTDP-beta-L-rhamnose biosynthesis in ...
|
|
69266
|
Engineered Bacillus pumilus laccase-like multi- copper oxidase for enhanced oxidation of the lignin model ...
|
|
69267
|
Engineered Bacillus pumilus laccase-like multi- copper oxidase for enhanced oxidation of the lignin model ...
|
|
69268
|
Engineered Bacillus pumilus laccase-like multi- copper oxidase for enhanced oxidation of the lignin model ...
|
|
69269
|
Engineered Bacillus pumilus laccase-like multi- copper oxidase for enhanced oxidation of the lignin model ...
|
|
69270
|
Engineered Bacillus pumilus laccase-like multi- copper oxidase for enhanced oxidation of the lignin model ...
|
|
69271
|
Engineered Bacillus pumilus laccase-like multi- copper oxidase for enhanced oxidation of the lignin model ...
|
|
69272
|
Engineered Bacillus pumilus laccase-like multi- copper oxidase for enhanced oxidation of the lignin model ...
|
|
69273
|
Engineered Bacillus pumilus laccase-like multi- copper oxidase for enhanced oxidation of the lignin model ...
|
|
69274
|
Engineered Bacillus pumilus laccase-like multi- copper oxidase for enhanced oxidation of the lignin model ...
|
|
69275
|
Inhibition of highly purified mammalian phospholipases A2 by non-steroidal anti-inflammatory agents. ...
|
|
69276
|
Inhibition of highly purified mammalian phospholipases A2 by non-steroidal anti-inflammatory agents. ...
|
|
69277
|
Inhibition of highly purified mammalian phospholipases A2 by non-steroidal anti-inflammatory agents. ...
|
|
69278
|
Inhibition of highly purified mammalian phospholipases A2 by non-steroidal anti-inflammatory agents. ...
|
|
69279
|
Inhibition of highly purified mammalian phospholipases A2 by non-steroidal anti-inflammatory agents. ...
|
|
69280
|
Inhibition of highly purified mammalian phospholipases A2 by non-steroidal anti-inflammatory agents. ...
|
|
69281
|
Inhibition of highly purified mammalian phospholipases A2 by non-steroidal anti-inflammatory agents. ...
|
|
69282
|
The first acidobacterial laccase-like multicopper oxidase revealed by metagenomics shows high salt and ...
|
|
69283
|
The first acidobacterial laccase-like multicopper oxidase revealed by metagenomics shows high salt and ...
|
|
69284
|
The first acidobacterial laccase-like multicopper oxidase revealed by metagenomics shows high salt and ...
|
|
69285
|
Overexpression of a Laccase with Dye Decolorization Activity from Bacillus sp. Induced in Escherichia coli
|
|
69286
|
Overexpression of a Laccase with Dye Decolorization Activity from Bacillus sp. Induced in Escherichia coli
|
|
69287
|
Overexpression of a Laccase with Dye Decolorization Activity from Bacillus sp. Induced in Escherichia coli
|
|
69288
|
Characterization of a mildly alkalophilic and thermostable recombinant Thermus thermophilus laccase with ...
|
|
69289
|
Characterization of a mildly alkalophilic and thermostable recombinant Thermus thermophilus laccase with ...
|
|
69290
|
Characterization of a mildly alkalophilic and thermostable recombinant Thermus thermophilus laccase with ...
|
|
69291
|
Characterization of a mildly alkalophilic and thermostable recombinant Thermus thermophilus laccase with ...
|
|
69292
|
Characterization of a mildly alkalophilic and thermostable recombinant Thermus thermophilus laccase with ...
|
|
69293
|
Characterization of a mildly alkalophilic and thermostable recombinant Thermus thermophilus laccase with ...
|
|
69294
|
Secretory expression of recombinant small laccase from Streptomyces coelicolor A3(2) in Pichia pastoris
|
|
69295
|
Secretory expression of recombinant small laccase from Streptomyces coelicolor A3(2) in Pichia pastoris
|
|
69296
|
Secretory expression of recombinant small laccase from Streptomyces coelicolor A3(2) in Pichia pastoris
|
|
69297
|
Functional expression enhancement of Bacillus pumilus CotA-laccase mutant WLF through site- directed ...
|
|
69298
|
Functional expression enhancement of Bacillus pumilus CotA-laccase mutant WLF through site- directed ...
|
|
69299
|
Functional expression enhancement of Bacillus pumilus CotA-laccase mutant WLF through site- directed ...
|
|
69300
|
Functional expression enhancement of Bacillus pumilus CotA-laccase mutant WLF through site- directed ...
|
|
69301
|
Functional expression enhancement of Bacillus pumilus CotA-laccase mutant WLF through site- directed ...
|
|
69302
|
Catalytic activation of Bacillus laccase after temperature treatment: Structural & biochemical ...
|
|
69303
|
Catalytic activation of Bacillus laccase after temperature treatment: Structural & biochemical ...
|
|
69304
|
Activity enhancement of CotA laccase by hydrophilic engineering, histidine tag optimization and static culture
|
|
69305
|
Activity enhancement of CotA laccase by hydrophilic engineering, histidine tag optimization and static culture
|
|
69306
|
Activity enhancement of CotA laccase by hydrophilic engineering, histidine tag optimization and static culture
|
|
69307
|
Activity enhancement of CotA laccase by hydrophilic engineering, histidine tag optimization and static culture
|
|
69308
|
Activity enhancement of CotA laccase by hydrophilic engineering, histidine tag optimization and static culture
|
|
69309
|
Activity enhancement of CotA laccase by hydrophilic engineering, histidine tag optimization and static culture
|
|
69310
|
Activity enhancement of CotA laccase by hydrophilic engineering, histidine tag optimization and static culture
|
|
69311
|
The development of CotA mediator cocktail system for dyes decolorization
|
|
69312
|
Rational design toward developing a more efficient laccase: Catalytic efficiency and selectivity
|
|
69313
|
Rational design toward developing a more efficient laccase: Catalytic efficiency and selectivity
|
|
69314
|
Rational design toward developing a more efficient laccase: Catalytic efficiency and selectivity
|
|
69315
|
Rational design toward developing a more efficient laccase: Catalytic efficiency and selectivity
|
|
69316
|
Rational design toward developing a more efficient laccase: Catalytic efficiency and selectivity
|
|
69317
|
Rational design toward developing a more efficient laccase: Catalytic efficiency and selectivity
|
|
69318
|
Rational design toward developing a more efficient laccase: Catalytic efficiency and selectivity
|
|
69319
|
Rational design toward developing a more efficient laccase: Catalytic efficiency and selectivity
|
|
69320
|
Rational design toward developing a more efficient laccase: Catalytic efficiency and selectivity
|
|
69321
|
Rational design toward developing a more efficient laccase: Catalytic efficiency and selectivity
|
|
69322
|
Rational design toward developing a more efficient laccase: Catalytic efficiency and selectivity
|
|
69323
|
Rational design toward developing a more efficient laccase: Catalytic efficiency and selectivity
|
|
69324
|
Rational design toward developing a more efficient laccase: Catalytic efficiency and selectivity
|
|
69325
|
Rational design toward developing a more efficient laccase: Catalytic efficiency and selectivity
|
|
69326
|
Rational design toward developing a more efficient laccase: Catalytic efficiency and selectivity
|
|
69327
|
Rational design toward developing a more efficient laccase: Catalytic efficiency and selectivity
|
|
69328
|
Four second-sphere residues of Thermus thermophilus SG0.5JP17-16 laccase tune the catalysis by ...
|
|
69329
|
Four second-sphere residues of Thermus thermophilus SG0.5JP17-16 laccase tune the catalysis by ...
|
|
69330
|
Four second-sphere residues of Thermus thermophilus SG0.5JP17-16 laccase tune the catalysis by ...
|
|
69331
|
Four second-sphere residues of Thermus thermophilus SG0.5JP17-16 laccase tune the catalysis by ...
|
|
69332
|
Four second-sphere residues of Thermus thermophilus SG0.5JP17-16 laccase tune the catalysis by ...
|
|
69333
|
Four second-sphere residues of Thermus thermophilus SG0.5JP17-16 laccase tune the catalysis by ...
|
|
69334
|
Four second-sphere residues of Thermus thermophilus SG0.5JP17-16 laccase tune the catalysis by ...
|
|
69335
|
Four second-sphere residues of Thermus thermophilus SG0.5JP17-16 laccase tune the catalysis by ...
|
|
69336
|
Four second-sphere residues of Thermus thermophilus SG0.5JP17-16 laccase tune the catalysis by ...
|
|
69337
|
Gene cloning, identification, and characterization of the multicopper oxidase CumA from Pseudomonas sp. 593
|
|
69338
|
Gene cloning, identification, and characterization of the multicopper oxidase CumA from Pseudomonas sp. 593
|
|
69339
|
Gene cloning, identification, and characterization of the multicopper oxidase CumA from Pseudomonas sp. 593
|
|
69340
|
Crystal structure of CotA laccase complexed with 2,2-azinobis-(3-ethylbenzothiazoline-6-sulfonate) at a novel ...
|
|
69341
|
Crystal structure of CotA laccase complexed with 2,2-azinobis-(3-ethylbenzothiazoline-6-sulfonate) at a novel ...
|
|
69342
|
Crystal structure of CotA laccase complexed with 2,2-azinobis-(3-ethylbenzothiazoline-6-sulfonate) at a novel ...
|
|
69343
|
Crystal structure of CotA laccase complexed with 2,2-azinobis-(3-ethylbenzothiazoline-6-sulfonate) at a novel ...
|
|
69344
|
Crystal structure of CotA laccase complexed with 2,2-azinobis-(3-ethylbenzothiazoline-6-sulfonate) at a novel ...
|
|
69345
|
Crystal structure of CotA laccase complexed with 2,2-azinobis-(3-ethylbenzothiazoline-6-sulfonate) at a novel ...
|
|
69346
|
Crystal structure of CotA laccase complexed with 2,2-azinobis-(3-ethylbenzothiazoline-6-sulfonate) at a novel ...
|
|
69347
|
Crystal structure of CotA laccase complexed with 2,2-azinobis-(3-ethylbenzothiazoline-6-sulfonate) at a novel ...
|
|
69348
|
Crystal structure of CotA laccase complexed with 2,2-azinobis-(3-ethylbenzothiazoline-6-sulfonate) at a novel ...
|
|
69349
|
Crystal structure of CotA laccase complexed with 2,2-azinobis-(3-ethylbenzothiazoline-6-sulfonate) at a novel ...
|
|
69350
|
Crystal structure of CotA laccase complexed with 2,2-azinobis-(3-ethylbenzothiazoline-6-sulfonate) at a novel ...
|
|
69351
|
Crystal structure of CotA laccase complexed with 2,2-azinobis-(3-ethylbenzothiazoline-6-sulfonate) at a novel ...
|
|
69352
|
Crystal structure of CotA laccase complexed with 2,2-azinobis-(3-ethylbenzothiazoline-6-sulfonate) at a novel ...
|
|
69353
|
Crystal structure of CotA laccase complexed with 2,2-azinobis-(3-ethylbenzothiazoline-6-sulfonate) at a novel ...
|
|
69354
|
Crystal structure of CotA laccase complexed with 2,2-azinobis-(3-ethylbenzothiazoline-6-sulfonate) at a novel ...
|
|
69355
|
Crystal structure of CotA laccase complexed with 2,2-azinobis-(3-ethylbenzothiazoline-6-sulfonate) at a novel ...
|
|
69356
|
Characterization of a Highly Thermostable and Organic Solvent-Tolerant Copper-Containing Polyphenol Oxidase ...
|
|
69357
|
Characterization of a Highly Thermostable and Organic Solvent-Tolerant Copper-Containing Polyphenol Oxidase ...
|
|
69358
|
In-Vitro Refolding and Characterization of Recombinant Laccase (CotA) From Bacillus pumilus MK001 and Its ...
|
|
69359
|
In-Vitro Refolding and Characterization of Recombinant Laccase (CotA) From Bacillus pumilus MK001 and Its ...
|
|
69360
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69361
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69362
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69363
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69364
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69365
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69366
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69367
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69368
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69369
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69370
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69371
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69372
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69373
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69374
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69375
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69376
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69377
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69378
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69379
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69380
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69381
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69382
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69383
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69384
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69385
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69386
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69387
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69388
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69389
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69390
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69391
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69392
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69393
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69394
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69395
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69396
|
Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate ...
|
|
69397
|
Cryptosporidium parvum spermidine/spermine N1-acetyltransferase exhibits different characteristics from the ...
|
|
69398
|
Cryptosporidium parvum spermidine/spermine N1-acetyltransferase exhibits different characteristics from the ...
|
|
69399
|
Cryptosporidium parvum spermidine/spermine N1-acetyltransferase exhibits different characteristics from the ...
|
|
69400
|
Cryptosporidium parvum spermidine/spermine N1-acetyltransferase exhibits different characteristics from the ...
|
|
69401
|
Cryptosporidium parvum spermidine/spermine N1-acetyltransferase exhibits different characteristics from the ...
|
|
69402
|
Cryptosporidium parvum spermidine/spermine N1-acetyltransferase exhibits different characteristics from the ...
|
|
69403
|
Cryptosporidium parvum spermidine/spermine N1-acetyltransferase exhibits different characteristics from the ...
|
|
69404
|
Cryptosporidium parvum spermidine/spermine N1-acetyltransferase exhibits different characteristics from the ...
|
|
69405
|
Cryptosporidium parvum spermidine/spermine N1-acetyltransferase exhibits different characteristics from the ...
|
|
69406
|
Cryptosporidium parvum spermidine/spermine N1-acetyltransferase exhibits different characteristics from the ...
|
|
69407
|
Structural and redox properties of the small laccase Ssl1 from Streptomyces sviceus.
|
|
69408
|
Structural and redox properties of the small laccase Ssl1 from Streptomyces sviceus.
|
|
69409
|
Structural and redox properties of the small laccase Ssl1 from Streptomyces sviceus.
|
|
69410
|
Structural and redox properties of the small laccase Ssl1 from Streptomyces sviceus.
|
|
69411
|
Structural and redox properties of the small laccase Ssl1 from Streptomyces sviceus.
|
|
69412
|
Structural and redox properties of the small laccase Ssl1 from Streptomyces sviceus.
|
|
69413
|
Expression, refolding, and characterization of a small laccase from Thermus thermophilus HJ6
|
|
69414
|
Expression, refolding, and characterization of a small laccase from Thermus thermophilus HJ6
|
|
69415
|
Expression, refolding, and characterization of a small laccase from Thermus thermophilus HJ6
|
|
69416
|
Expression, refolding, and characterization of a small laccase from Thermus thermophilus HJ6
|
|
69417
|
Expression, refolding, and characterization of a small laccase from Thermus thermophilus HJ6
|
|
69418
|
Expression, refolding, and characterization of a small laccase from Thermus thermophilus HJ6
|
|
69419
|
Expression, refolding, and characterization of a small laccase from Thermus thermophilus HJ6
|
|
69420
|
Expression, refolding, and characterization of a small laccase from Thermus thermophilus HJ6
|
|
69421
|
Improving the catalytic efficiency of Bacillus pumilus CotA-laccase by site-directed mutagenesis
|
|
69422
|
Improving the catalytic efficiency of Bacillus pumilus CotA-laccase by site-directed mutagenesis
|
|
69423
|
Improving the catalytic efficiency of Bacillus pumilus CotA-laccase by site-directed mutagenesis
|
|
69424
|
Improving the catalytic efficiency of Bacillus pumilus CotA-laccase by site-directed mutagenesis
|
|
69425
|
Improving the catalytic efficiency of Bacillus pumilus CotA-laccase by site-directed mutagenesis
|
|
69426
|
Improving the catalytic efficiency of Bacillus pumilus CotA-laccase by site-directed mutagenesis
|
|
69427
|
Improving the catalytic efficiency of Bacillus pumilus CotA-laccase by site-directed mutagenesis
|
|
69428
|
Improving the catalytic efficiency of Bacillus pumilus CotA-laccase by site-directed mutagenesis
|
|
69429
|
Improving the catalytic efficiency of Bacillus pumilus CotA-laccase by site-directed mutagenesis
|
|
69430
|
Improving the catalytic efficiency of Bacillus pumilus CotA-laccase by site-directed mutagenesis
|
|
69431
|
Improving the catalytic efficiency of Bacillus pumilus CotA-laccase by site-directed mutagenesis
|
|
69432
|
Improving the catalytic efficiency of Bacillus pumilus CotA-laccase by site-directed mutagenesis
|
|
69433
|
Optimization of a small laccase by active-site redesign
|
|
69434
|
Optimization of a small laccase by active-site redesign
|
|
69435
|
Optimization of a small laccase by active-site redesign
|
|
69436
|
Optimization of a small laccase by active-site redesign
|
|
69437
|
Optimization of a small laccase by active-site redesign
|
|
69438
|
Optimization of a small laccase by active-site redesign
|
|
69439
|
Optimization of a small laccase by active-site redesign
|
|
69440
|
Optimization of a small laccase by active-site redesign
|
|
69441
|
Biochemical properties and yields of diverse bacterial laccase-like multicopper oxidases expressed in ...
|
|
69442
|
Biochemical properties and yields of diverse bacterial laccase-like multicopper oxidases expressed in ...
|
|
69443
|
Biochemical properties and yields of diverse bacterial laccase-like multicopper oxidases expressed in ...
|
|
69444
|
Biochemical properties and yields of diverse bacterial laccase-like multicopper oxidases expressed in ...
|
|
69445
|
A semi-rational approach to obtain an ionic liquid tolerant bacterial laccase through π-type interactions
|
|
69446
|
A semi-rational approach to obtain an ionic liquid tolerant bacterial laccase through π-type interactions
|
|
69447
|
A semi-rational approach to obtain an ionic liquid tolerant bacterial laccase through π-type interactions
|
|
69448
|
A semi-rational approach to obtain an ionic liquid tolerant bacterial laccase through π-type interactions
|
|
69449
|
A semi-rational approach to obtain an ionic liquid tolerant bacterial laccase through π-type interactions
|
|
69450
|
A semi-rational approach to obtain an ionic liquid tolerant bacterial laccase through π-type interactions
|
|
69451
|
A semi-rational approach to obtain an ionic liquid tolerant bacterial laccase through π-type interactions
|
|
69452
|
A semi-rational approach to obtain an ionic liquid tolerant bacterial laccase through π-type interactions
|
|
69453
|
A semi-rational approach to obtain an ionic liquid tolerant bacterial laccase through π-type interactions
|
|
69454
|
A semi-rational approach to obtain an ionic liquid tolerant bacterial laccase through π-type interactions
|
|
69455
|
A semi-rational approach to obtain an ionic liquid tolerant bacterial laccase through π-type interactions
|
|
69456
|
A semi-rational approach to obtain an ionic liquid tolerant bacterial laccase through π-type interactions
|
|
69457
|
A semi-rational approach to obtain an ionic liquid tolerant bacterial laccase through π-type interactions
|
|
69458
|
A 24.7-kDa copper- containing oxidase, secreted by Thermobifida fusca, significantly increasing the ...
|
|
69459
|
Characterization of the alkaline laccase Ssl1 from Streptomyces sviceus with unusual properties discovered by ...
|
|
69460
|
Characterization of the alkaline laccase Ssl1 from Streptomyces sviceus with unusual properties discovered by ...
|
|
69461
|
Characterization of the alkaline laccase Ssl1 from Streptomyces sviceus with unusual properties discovered by ...
|
|
69462
|
Purification and characterization of an extracellular, thermo-alkali-stable, metal tolerant laccase from ...
|
|
69463
|
Purification and characterization of an extracellular, thermo-alkali-stable, metal tolerant laccase from ...
|
|
69464
|
Purification and characterization of an extracellular, thermo-alkali-stable, metal tolerant laccase from ...
|
|
69465
|
Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required for copper ...
|
|
69466
|
A thermostable laccase from Streptomyces lavendulae REN-7: purification, characterization, nucleotide ...
|
|
69467
|
A thermostable laccase from Streptomyces lavendulae REN-7: purification, characterization, nucleotide ...
|
|
69468
|
A thermostable laccase from Streptomyces lavendulae REN-7: purification, characterization, nucleotide ...
|
|
69469
|
A thermostable laccase from Streptomyces lavendulae REN-7: purification, characterization, nucleotide ...
|
|
69470
|
Cloning and expression of thermo-alkali-stable laccase of Bacillus licheniformis in Pichia pastoris and its ...
|
|
69471
|
Cloning and expression of thermo-alkali-stable laccase of Bacillus licheniformis in Pichia pastoris and its ...
|
|
69472
|
Cloning and expression of thermo-alkali-stable laccase of Bacillus licheniformis in Pichia pastoris and its ...
|
|
69473
|
Alkali and halide-resistant catalysis by the multipotent oxidase from Marinomonas mediterranea
|
|
69474
|
Alkali and halide-resistant catalysis by the multipotent oxidase from Marinomonas mediterranea
|
|
69475
|
Alkali and halide-resistant catalysis by the multipotent oxidase from Marinomonas mediterranea
|
|
69476
|
Alkali and halide-resistant catalysis by the multipotent oxidase from Marinomonas mediterranea
|
|
69477
|
Alkali and halide-resistant catalysis by the multipotent oxidase from Marinomonas mediterranea
|
|
69478
|
Alkali and halide-resistant catalysis by the multipotent oxidase from Marinomonas mediterranea
|
|
69479
|
Alkali and halide-resistant catalysis by the multipotent oxidase from Marinomonas mediterranea
|
|
69480
|
Alkali and halide-resistant catalysis by the multipotent oxidase from Marinomonas mediterranea
|
|
69481
|
Alkali and halide-resistant catalysis by the multipotent oxidase from Marinomonas mediterranea
|
|
69482
|
Alkali and halide-resistant catalysis by the multipotent oxidase from Marinomonas mediterranea
|
|
69483
|
Alkali and halide-resistant catalysis by the multipotent oxidase from Marinomonas mediterranea
|
|
69484
|
The multicopper oxidase CutO confers copper tolerance to Rhodobacter capsulatus
|
|
69485
|
The multicopper oxidase CutO confers copper tolerance to Rhodobacter capsulatus
|
|
69486
|
The multicopper oxidase CutO confers copper tolerance to Rhodobacter capsulatus
|
|
69487
|
The multicopper oxidase CutO confers copper tolerance to Rhodobacter capsulatus
|
|
69488
|
Identification of a novel copper-activated and halide-tolerant laccase in Geobacillus thermopakistaniensis
|
|
69489
|
Identification of a novel copper-activated and halide-tolerant laccase in Geobacillus thermopakistaniensis
|
|
69490
|
Heterologous expression of a Streptomyces cyaneus laccase for biomass modification applications
|
|
69491
|
Heterologous expression of a Streptomyces cyaneus laccase for biomass modification applications
|
|
69492
|
Heterologous expression of a Streptomyces cyaneus laccase for biomass modification applications
|
|
69493
|
Purification and characterization of a thermostable laccase with unique oxidative characteristics from ...
|
|
69494
|
Purification and characterization of a thermostable laccase with unique oxidative characteristics from ...
|
|
69495
|
Cloning and characterization of a new laccase from Lactobacillus plantarum J16 CECT 8944 catalyzing biogenic ...
|
|
69496
|
Cloning and characterization of a new laccase from Lactobacillus plantarum J16 CECT 8944 catalyzing biogenic ...
|
|
69497
|
Directed evolution of CotA laccase for increased substrate specificity using Bacillus subtilis spores
|
|
69498
|
Directed evolution of CotA laccase for increased substrate specificity using Bacillus subtilis spores
|
|
69499
|
Directed evolution of CotA laccase for increased substrate specificity using Bacillus subtilis spores
|
|
69500
|
Directed evolution of CotA laccase for increased substrate specificity using Bacillus subtilis spores
|
|
69501
|
An efficient in vitro refolding of recombinant bacterial laccase in Escherichia coli.
|
|
69502
|
An efficient in vitro refolding of recombinant bacterial laccase in Escherichia coli.
|
|
69503
|
Enhancement of catalysis and functional expression of a bacterial laccase by single amino acid replacement.
|
|
69504
|
Enhancement of catalysis and functional expression of a bacterial laccase by single amino acid replacement.
|
|
69505
|
Enhancement of catalysis and functional expression of a bacterial laccase by single amino acid replacement.
|
|
69506
|
Enhancement of catalysis and functional expression of a bacterial laccase by single amino acid replacement.
|
|
69507
|
Enhancement of catalysis and functional expression of a bacterial laccase by single amino acid replacement.
|
|
69508
|
Enhancement of catalysis and functional expression of a bacterial laccase by single amino acid replacement.
|
|
69509
|
Enhancement of catalysis and functional expression of a bacterial laccase by single amino acid replacement.
|
|
69510
|
Enhancement of catalysis and functional expression of a bacterial laccase by single amino acid replacement.
|
|
69511
|
Enhancement of catalysis and functional expression of a bacterial laccase by single amino acid replacement.
|
|
69512
|
Enhancement of catalysis and functional expression of a bacterial laccase by single amino acid replacement.
|
|
69513
|
Enhancement of catalysis and functional expression of a bacterial laccase by single amino acid replacement.
|
|
69514
|
CotA, a multicopper oxidase from Bacillus pumilus WH4, exhibits manganese-oxidase activity.
|
|
69515
|
CotA, a multicopper oxidase from Bacillus pumilus WH4, exhibits manganese-oxidase activity.
|
|
69516
|
Involvement of Tyr108 in the enzyme mechanism of the small laccase from Streptomyces coelicolor.
|
|
69517
|
Involvement of Tyr108 in the enzyme mechanism of the small laccase from Streptomyces coelicolor.
|
|
69518
|
Involvement of Tyr108 in the enzyme mechanism of the small laccase from Streptomyces coelicolor.
|
|
69519
|
Involvement of Tyr108 in the enzyme mechanism of the small laccase from Streptomyces coelicolor.
|
|
69520
|
Involvement of Tyr108 in the enzyme mechanism of the small laccase from Streptomyces coelicolor.
|
|
69521
|
Involvement of Tyr108 in the enzyme mechanism of the small laccase from Streptomyces coelicolor.
|
|
69522
|
A new multicopper oxidase from Gram- positive bacterium Rhodococcus erythropolis with activity modulating ...
|
|
69523
|
A new multicopper oxidase from Gram- positive bacterium Rhodococcus erythropolis with activity modulating ...
|
|
69524
|
A new multicopper oxidase from Gram- positive bacterium Rhodococcus erythropolis with activity modulating ...
|
|
69525
|
A new multicopper oxidase from Gram- positive bacterium Rhodococcus erythropolis with activity modulating ...
|
|
69526
|
A new multicopper oxidase from Gram- positive bacterium Rhodococcus erythropolis with activity modulating ...
|
|
69527
|
A new multicopper oxidase from Gram- positive bacterium Rhodococcus erythropolis with activity modulating ...
|
|
69528
|
A new multicopper oxidase from Gram- positive bacterium Rhodococcus erythropolis with activity modulating ...
|
|
69529
|
A new multicopper oxidase from Gram- positive bacterium Rhodococcus erythropolis with activity modulating ...
|
|
69530
|
A new multicopper oxidase from Gram- positive bacterium Rhodococcus erythropolis with activity modulating ...
|
|
69531
|
A new multicopper oxidase from Gram- positive bacterium Rhodococcus erythropolis with activity modulating ...
|
|
69532
|
A new multicopper oxidase from Gram- positive bacterium Rhodococcus erythropolis with activity modulating ...
|
|
69533
|
A new multicopper oxidase from Gram- positive bacterium Rhodococcus erythropolis with activity modulating ...
|
|
69534
|
A new multicopper oxidase from Gram- positive bacterium Rhodococcus erythropolis with activity modulating ...
|
|
69535
|
A new multicopper oxidase from Gram- positive bacterium Rhodococcus erythropolis with activity modulating ...
|
|
69536
|
Biochemical characterization of a thermostable cobalt- or copper-dependent polyphenol oxidase with dye ...
|
|
69537
|
Biochemical characterization of a thermostable cobalt- or copper-dependent polyphenol oxidase with dye ...
|
|
69538
|
Biochemical characterization of a thermostable cobalt- or copper-dependent polyphenol oxidase with dye ...
|
|
69539
|
Biochemical characterization of a thermostable cobalt- or copper-dependent polyphenol oxidase with dye ...
|
|
69540
|
Production of polyextremotolerant laccase by Achromobacter xylosoxidans HWN16 and Citrobacter ...
|
|
69541
|
Production of polyextremotolerant laccase by Achromobacter xylosoxidans HWN16 and Citrobacter ...
|
|
69542
|
Engineering CotA Laccase for Acidic pH Stability Using Bacillus subtilis Spore Display
|
|
69543
|
Engineering CotA Laccase for Acidic pH Stability Using Bacillus subtilis Spore Display
|
|
69544
|
Engineering CotA Laccase for Acidic pH Stability Using Bacillus subtilis Spore Display
|
|
69545
|
Engineering CotA Laccase for Acidic pH Stability Using Bacillus subtilis Spore Display
|
|
69546
|
Engineering CotA Laccase for Acidic pH Stability Using Bacillus subtilis Spore Display
|
|
69547
|
Engineering CotA Laccase for Acidic pH Stability Using Bacillus subtilis Spore Display
|
|
69548
|
Engineering CotA Laccase for Acidic pH Stability Using Bacillus subtilis Spore Display
|
|
69549
|
Enhanced expression of a recombinant bacterial laccase at low temperature and microaerobic conditions: ...
|
|
69550
|
Enhanced expression of a recombinant bacterial laccase at low temperature and microaerobic conditions: ...
|
|
69551
|
Enhanced expression of a recombinant bacterial laccase at low temperature and microaerobic conditions: ...
|
|
69552
|
Enhanced expression of a recombinant bacterial laccase at low temperature and microaerobic conditions: ...
|
|
69553
|
Serine hydroxymethyltransferase from Escherichia coli: purification and properties
|
|
69554
|
Serine hydroxymethyltransferase from Escherichia coli: purification and properties
|
|
69555
|
Serine hydroxymethyltransferase from Escherichia coli: purification and properties
|
|
69556
|
Serine hydroxymethyltransferase from Escherichia coli: purification and properties
|
|
69557
|
Serine hydroxymethyltransferase from Escherichia coli: purification and properties
|
|
69558
|
Serine hydroxymethyltransferase from Escherichia coli: purification and properties
|
|
69559
|
Serine hydroxymethyltransferase from Escherichia coli: purification and properties
|
|
69560
|
Serine hydroxymethyltransferase from Escherichia coli: purification and properties
|
|
69561
|
Serine hydroxymethyltransferase from Escherichia coli: purification and properties
|
|
69562
|
Methyl syringate: an efficient phenolic mediator for bacterial and fungal laccases.
|
|
69563
|
Methyl syringate: an efficient phenolic mediator for bacterial and fungal laccases.
|
|
69564
|
Methyl syringate: an efficient phenolic mediator for bacterial and fungal laccases.
|
|
69565
|
Methyl syringate: an efficient phenolic mediator for bacterial and fungal laccases.
|
|
69566
|
Methyl syringate: an efficient phenolic mediator for bacterial and fungal laccases.
|
|
69567
|
Methyl syringate: an efficient phenolic mediator for bacterial and fungal laccases.
|
|
69568
|
Plackett-Burman Design for rGILCC1 Laccase Activity Enhancement in Pichia pastoris: Concentrated Enzyme ...
|
|
69569
|
Process optimization, purification and characterization of alkaline stable white laccase from Myrothecium ...
|
|
69570
|
A novel white laccase from Pleurotus ostreatus
|
|
69571
|
A novel white laccase from Pleurotus ostreatus
|
|
69572
|
A novel white laccase from Pleurotus ostreatus
|
|
69573
|
A novel white laccase from Pleurotus ostreatus
|
|
69574
|
A novel white laccase from Pleurotus ostreatus
|
|
69575
|
A novel white laccase from Pleurotus ostreatus
|
|
69576
|
A novel white laccase from Pleurotus ostreatus
|
|
69577
|
A novel white laccase from Pleurotus ostreatus
|
|
69578
|
A novel white laccase from Pleurotus ostreatus
|
|
69579
|
A novel white laccase from Pleurotus ostreatus
|
|
69580
|
A novel white laccase from Pleurotus ostreatus
|
|
69581
|
A novel white laccase from Pleurotus ostreatus
|
|
69582
|
A novel white laccase from Pleurotus ostreatus
|
|
69583
|
A novel white laccase from Pleurotus ostreatus
|
|
69584
|
A novel white laccase from Pleurotus ostreatus
|
|
69585
|
A novel white laccase from Pleurotus ostreatus
|
|
69586
|
A novel white laccase from Pleurotus ostreatus
|
|
69587
|
Characterization of C-terminally engineered laccases.
|
|
69588
|
Characterization of C-terminally engineered laccases.
|
|
69589
|
Characterization of C-terminally engineered laccases.
|
|
69590
|
Characterization of C-terminally engineered laccases.
|
|
69591
|
Characterization of C-terminally engineered laccases.
|
|
69592
|
Characterization of C-terminally engineered laccases.
|
|
69593
|
Characterization of C-terminally engineered laccases.
|
|
69594
|
Characterization of C-terminally engineered laccases.
|
|
69595
|
Characterization of C-terminally engineered laccases.
|
|
69596
|
Characterization of C-terminally engineered laccases.
|
|
69597
|
Characterization of C-terminally engineered laccases.
|
|
69598
|
Characterization of C-terminally engineered laccases.
|
|
69599
|
Enhanced catalytic efficiency of CotA-laccase by DNA shuffling
|
|
69600
|
Enhanced catalytic efficiency of CotA-laccase by DNA shuffling
|
|
69601
|
Isolation and characterization of an alkali and thermostable laccase from a novel Alcaligenes faecalis ...
|
|
69602
|
Isolation and characterization of an alkali and thermostable laccase from a novel Alcaligenes faecalis ...
|
|
69603
|
Isolation and characterization of an alkali and thermostable laccase from a novel Alcaligenes faecalis ...
|
|
69604
|
Bacillus pumilus laccase: a heat stable enzyme with a wide substrate spectrum.
|
|
69605
|
Bacillus pumilus laccase: a heat stable enzyme with a wide substrate spectrum.
|
|
69606
|
Bacillus pumilus laccase: a heat stable enzyme with a wide substrate spectrum.
|
|
69607
|
Bacillus pumilus laccase: a heat stable enzyme with a wide substrate spectrum.
|
|
69608
|
Bacillus pumilus laccase: a heat stable enzyme with a wide substrate spectrum.
|
|
69609
|
A strategy for soluble overexpression and biochemical characterization of halo-thermotolerant Bacillus laccase ...
|
|
69610
|
A strategy for soluble overexpression and biochemical characterization of halo-thermotolerant Bacillus laccase ...
|
|
69611
|
Laccase catalysed oxidation of syringic acid: calorimetric determination of kinetic parameters.
|
|
69612
|
Laccase catalysed oxidation of syringic acid: calorimetric determination of kinetic parameters.
|
|
69613
|
Blue laccase from Galerina sp.: Properties and potential for Kraft lignin demethylation
|
|
69614
|
Blue laccase from Galerina sp.: Properties and potential for Kraft lignin demethylation
|
|
69615
|
A novel fluorimetric method for laccase activities measurement using Amplex Red as substrate.
|
|
69616
|
Biochemical studies of the multicopper oxidase (small laccase) from Streptomyces coelicolor using bioactive ...
|
|
69617
|
Biochemical studies of the multicopper oxidase (small laccase) from Streptomyces coelicolor using bioactive ...
|
|
69618
|
Biochemical studies of the multicopper oxidase (small laccase) from Streptomyces coelicolor using bioactive ...
|
|
69619
|
Biochemical studies of the multicopper oxidase (small laccase) from Streptomyces coelicolor using bioactive ...
|
|
69620
|
Biochemical studies of the multicopper oxidase (small laccase) from Streptomyces coelicolor using bioactive ...
|
|
69621
|
Biochemical studies of the multicopper oxidase (small laccase) from Streptomyces coelicolor using bioactive ...
|
|
69622
|
Biochemical studies of the multicopper oxidase (small laccase) from Streptomyces coelicolor using bioactive ...
|
|
69623
|
Biochemical studies of the multicopper oxidase (small laccase) from Streptomyces coelicolor using bioactive ...
|
|
69624
|
Enhanced expression of an industry applicable CotA laccase from Bacillus subtilis in Pichia pastoris by ...
|
|
69625
|
Enhanced expression of an industry applicable CotA laccase from Bacillus subtilis in Pichia pastoris by ...
|
|
69626
|
Cloning, expression, and characterization of a thermostable and pH-stable laccase from Klebsiella pneumoniae ...
|
|
69627
|
Cloning, expression, and characterization of a thermostable and pH-stable laccase from Klebsiella pneumoniae ...
|
|
69628
|
Immobilization of CotA, an extremophilic laccase from Bacillus subtilis, on glassy carbon electrodes for ...
|
|
69629
|
Immobilization of CotA, an extremophilic laccase from Bacillus subtilis, on glassy carbon electrodes for ...
|
|
69630
|
Immobilization of CotA, an extremophilic laccase from Bacillus subtilis, on glassy carbon electrodes for ...
|
|
69631
|
Immobilization of CotA, an extremophilic laccase from Bacillus subtilis, on glassy carbon electrodes for ...
|
|
69632
|
A thermostable laccase from Thermus sp. 2.9 and its potential for delignification of Eucalyptus biomass
|
|
69633
|
A thermostable laccase from Thermus sp. 2.9 and its potential for delignification of Eucalyptus biomass
|
|
69634
|
Expression and characterization of a thermostable organic solvent-tolerant laccase from Bacillus licheniformis ...
|
|
69635
|
Expression and characterization of a thermostable organic solvent-tolerant laccase from Bacillus licheniformis ...
|
|
69636
|
Expression and characterization of a thermostable organic solvent-tolerant laccase from Bacillus licheniformis ...
|
|
69637
|
Expression and characterization of a thermostable organic solvent-tolerant laccase from Bacillus licheniformis ...
|
|
69638
|
Expression and characterization of a thermostable organic solvent-tolerant laccase from Bacillus licheniformis ...
|
|
69639
|
Expression and characterization of a thermostable organic solvent-tolerant laccase from Bacillus licheniformis ...
|
|
69640
|
Expression and characterization of a thermostable organic solvent-tolerant laccase from Bacillus licheniformis ...
|
|
69641
|
Expression and characterization of a thermostable organic solvent-tolerant laccase from Bacillus licheniformis ...
|
|
69642
|
Expression and characterization of a thermostable organic solvent-tolerant laccase from Bacillus licheniformis ...
|
|
69643
|
Expression and characterization of a thermostable organic solvent-tolerant laccase from Bacillus licheniformis ...
|
|
69644
|
Expression and characterization of a thermostable organic solvent-tolerant laccase from Bacillus licheniformis ...
|
|
69645
|
Expression and characterization of a thermostable organic solvent-tolerant laccase from Bacillus licheniformis ...
|
|
69646
|
Expression and characterization of a thermostable organic solvent-tolerant laccase from Bacillus licheniformis ...
|
|
69647
|
Expression and characterization of a thermostable organic solvent-tolerant laccase from Bacillus licheniformis ...
|
|
69648
|
Expression and characterization of a thermostable organic solvent-tolerant laccase from Bacillus licheniformis ...
|
|
69649
|
Expression and characterization of a thermostable organic solvent-tolerant laccase from Bacillus licheniformis ...
|
|
69650
|
Purification and characterization of an extracellular laccase from a Pseudomonas sp. LBC1 and its application ...
|
|
69651
|
Purification and characterization of an alkaline chloride-tolerant laccase from a halotolerant bacterium, ...
|
|
69652
|
Purification and characterization of an alkaline chloride-tolerant laccase from a halotolerant bacterium, ...
|
|
69653
|
Purification of a thermostable alkaline laccase from papaya (Carica papaya) using affinity chromatography
|
|
69654
|
Purification of a thermostable alkaline laccase from papaya (Carica papaya) using affinity chromatography
|
|
69655
|
Purification of a thermostable alkaline laccase from papaya (Carica papaya) using affinity chromatography
|
|
69656
|
Axial bonds at the T1 Cu site of Thermus thermophilus SG0.5JP17-16 laccase influence enzymatic properties
|
|
69657
|
Axial bonds at the T1 Cu site of Thermus thermophilus SG0.5JP17-16 laccase influence enzymatic properties
|
|
69658
|
Axial bonds at the T1 Cu site of Thermus thermophilus SG0.5JP17-16 laccase influence enzymatic properties
|
|
69659
|
Axial bonds at the T1 Cu site of Thermus thermophilus SG0.5JP17-16 laccase influence enzymatic properties
|
|
69660
|
Axial bonds at the T1 Cu site of Thermus thermophilus SG0.5JP17-16 laccase influence enzymatic properties
|
|
69661
|
Axial bonds at the T1 Cu site of Thermus thermophilus SG0.5JP17-16 laccase influence enzymatic properties
|
|
69662
|
Characterization of a Novel, Cold-Adapted, and Thermostable Laccase-Like Enzyme With High Tolerance for ...
|
|
69663
|
Characterization of a Novel, Cold-Adapted, and Thermostable Laccase-Like Enzyme With High Tolerance for ...
|
|
69664
|
Characterization of a Novel, Cold-Adapted, and Thermostable Laccase-Like Enzyme With High Tolerance for ...
|
|
69665
|
Characterization of a Novel, Cold-Adapted, and Thermostable Laccase-Like Enzyme With High Tolerance for ...
|
|
69666
|
Characterization of a Novel, Cold-Adapted, and Thermostable Laccase-Like Enzyme With High Tolerance for ...
|
|
69667
|
A new Stenotrophomonas maltophilia strain producing laccase. Use in decolorization of synthetics dyes
|
|
69668
|
A new Stenotrophomonas maltophilia strain producing laccase. Use in decolorization of synthetics dyes
|
|
69669
|
A new Stenotrophomonas maltophilia strain producing laccase. Use in decolorization of synthetics dyes
|
|
69670
|
Detoxification of azo dyes by a novel pH-versatile, salt-resistant laccase from Streptomyces ipomoea
|
|
69671
|
Detoxification of azo dyes by a novel pH-versatile, salt-resistant laccase from Streptomyces ipomoea
|
|
69672
|
Detoxification of azo dyes by a novel pH-versatile, salt-resistant laccase from Streptomyces ipomoea
|
|
69673
|
LccA, an archaeal laccase secreted as a highly stable glycoprotein into the extracellular medium by Haloferax ...
|
|
69674
|
LccA, an archaeal laccase secreted as a highly stable glycoprotein into the extracellular medium by Haloferax ...
|
|
69675
|
LccA, an archaeal laccase secreted as a highly stable glycoprotein into the extracellular medium by Haloferax ...
|
|
69676
|
LccA, an archaeal laccase secreted as a highly stable glycoprotein into the extracellular medium by Haloferax ...
|
|
69677
|
LccA, an archaeal laccase secreted as a highly stable glycoprotein into the extracellular medium by Haloferax ...
|
|
69678
|
Roles of small laccases from Streptomyces in lignin degradation
|
|
69679
|
Roles of small laccases from Streptomyces in lignin degradation
|
|
69680
|
Roles of small laccases from Streptomyces in lignin degradation
|
|
69681
|
Roles of small laccases from Streptomyces in lignin degradation
|
|
69682
|
Purification and characterization of the constitutive form of laccase from the basidiomycete Coriolus hirsutus ...
|
|
69683
|
Purification and characterization of the constitutive form of laccase from the basidiomycete Coriolus hirsutus ...
|
|
69684
|
Purification and characterization of the constitutive form of laccase from the basidiomycete Coriolus hirsutus ...
|
|
69685
|
Purification and characterization of the constitutive form of laccase from the basidiomycete Coriolus hirsutus ...
|
|
69686
|
Purification and characterization of the constitutive form of laccase from the basidiomycete Coriolus hirsutus ...
|
|
69687
|
Purification and characterization of the constitutive form of laccase from the basidiomycete Coriolus hirsutus ...
|
|
69688
|
Characterization of an alkali- and halide- resistant laccase expressed in E. coli: CotA from Bacillus clausii
|
|
69689
|
Characterization of an alkali- and halide- resistant laccase expressed in E. coli: CotA from Bacillus clausii
|
|
69690
|
The role of Glu498 in the dioxygen reactivity of CotA- laccase from Bacillus subtilis
|
|
69691
|
The role of Glu498 in the dioxygen reactivity of CotA- laccase from Bacillus subtilis
|
|
69692
|
The role of Glu498 in the dioxygen reactivity of CotA- laccase from Bacillus subtilis
|
|
69693
|
The role of Glu498 in the dioxygen reactivity of CotA- laccase from Bacillus subtilis
|
|
69694
|
The role of Glu498 in the dioxygen reactivity of CotA- laccase from Bacillus subtilis
|
|
69695
|
The role of Glu498 in the dioxygen reactivity of CotA- laccase from Bacillus subtilis
|
|
69696
|
The role of Glu498 in the dioxygen reactivity of CotA- laccase from Bacillus subtilis
|
|
69697
|
The role of Glu498 in the dioxygen reactivity of CotA- laccase from Bacillus subtilis
|
|
69698
|
The role of Glu498 in the dioxygen reactivity of CotA- laccase from Bacillus subtilis
|
|
69699
|
The role of Glu498 in the dioxygen reactivity of CotA- laccase from Bacillus subtilis
|
|
69700
|
The role of Glu498 in the dioxygen reactivity of CotA- laccase from Bacillus subtilis
|
|
69701
|
The role of Glu498 in the dioxygen reactivity of CotA- laccase from Bacillus subtilis
|
|
69702
|
The role of Glu498 in the dioxygen reactivity of CotA- laccase from Bacillus subtilis
|
|
69703
|
The role of Glu498 in the dioxygen reactivity of CotA- laccase from Bacillus subtilis
|
|
69704
|
The role of Glu498 in the dioxygen reactivity of CotA- laccase from Bacillus subtilis
|
|
69705
|
The role of Glu498 in the dioxygen reactivity of CotA- laccase from Bacillus subtilis
|
|
69706
|
Cloning, expression in Escherichia coli, and enzymatic properties of laccase from Aeromonas hydrophila WL-11
|
|
69707
|
Cloning, expression in Escherichia coli, and enzymatic properties of laccase from Aeromonas hydrophila WL-11
|
|
69708
|
Bilirubin oxidase from Bacillus pumilus: a promising enzyme for the elaboration of efficient cathodes in ...
|
|
69709
|
Bilirubin oxidase from Bacillus pumilus: a promising enzyme for the elaboration of efficient cathodes in ...
|
|
69710
|
Bilirubin oxidase from Bacillus pumilus: a promising enzyme for the elaboration of efficient cathodes in ...
|
|
69711
|
Bilirubin oxidase from Bacillus pumilus: a promising enzyme for the elaboration of efficient cathodes in ...
|
|
69712
|
Bilirubin oxidase from Bacillus pumilus: a promising enzyme for the elaboration of efficient cathodes in ...
|
|
69713
|
Enhancement of a bacterial laccase thermostability through directed mutagenesis of a surface loop
|
|
69714
|
Enhancement of a bacterial laccase thermostability through directed mutagenesis of a surface loop
|
|
69715
|
Enhancement of a bacterial laccase thermostability through directed mutagenesis of a surface loop
|
|
69716
|
Enhancement of a bacterial laccase thermostability through directed mutagenesis of a surface loop
|
|
69717
|
Engineering Klebsiella sp. 601 multicopper oxidase enhances the catalytic efficiency towards phenolic ...
|
|
69718
|
Engineering Klebsiella sp. 601 multicopper oxidase enhances the catalytic efficiency towards phenolic ...
|
|
69719
|
Engineering Klebsiella sp. 601 multicopper oxidase enhances the catalytic efficiency towards phenolic ...
|
|
69720
|
Engineering Klebsiella sp. 601 multicopper oxidase enhances the catalytic efficiency towards phenolic ...
|
|
69721
|
Engineering Klebsiella sp. 601 multicopper oxidase enhances the catalytic efficiency towards phenolic ...
|
|
69722
|
Engineering Klebsiella sp. 601 multicopper oxidase enhances the catalytic efficiency towards phenolic ...
|
|
69723
|
Engineering Klebsiella sp. 601 multicopper oxidase enhances the catalytic efficiency towards phenolic ...
|
|
69724
|
Engineering Klebsiella sp. 601 multicopper oxidase enhances the catalytic efficiency towards phenolic ...
|
|
69725
|
Engineering Klebsiella sp. 601 multicopper oxidase enhances the catalytic efficiency towards phenolic ...
|
|
69726
|
Crystal structure of the multicopper oxidase from the pathogenic bacterium Campylobacter jejuni CGUG11284: ...
|
|
69727
|
Crystal structure of the multicopper oxidase from the pathogenic bacterium Campylobacter jejuni CGUG11284: ...
|
|
69728
|
Crystal structure of the multicopper oxidase from the pathogenic bacterium Campylobacter jejuni CGUG11284: ...
|
|
69729
|
Crystal structure of the multicopper oxidase from the pathogenic bacterium Campylobacter jejuni CGUG11284: ...
|
|
69730
|
Crystal structure of the multicopper oxidase from the pathogenic bacterium Campylobacter jejuni CGUG11284: ...
|
|
69731
|
Crystal structure of the multicopper oxidase from the pathogenic bacterium Campylobacter jejuni CGUG11284: ...
|
|
69732
|
Expression and Characterization of a Recombinant Laccase with Alkalistable and Thermostable Properties from ...
|
|
69733
|
Expression and Characterization of a Recombinant Laccase with Alkalistable and Thermostable Properties from ...
|
|
69734
|
Expression and Characterization of a Recombinant Laccase with Alkalistable and Thermostable Properties from ...
|
|
69735
|
The effect of mutations near the T1 copper site on the biochemical characteristics of the small laccase from ...
|
|
69736
|
The effect of mutations near the T1 copper site on the biochemical characteristics of the small laccase from ...
|
|
69737
|
The effect of mutations near the T1 copper site on the biochemical characteristics of the small laccase from ...
|
|
69738
|
The effect of mutations near the T1 copper site on the biochemical characteristics of the small laccase from ...
|
|
69739
|
The effect of mutations near the T1 copper site on the biochemical characteristics of the small laccase from ...
|
|
69740
|
The effect of mutations near the T1 copper site on the biochemical characteristics of the small laccase from ...
|
|
69741
|
The effect of mutations near the T1 copper site on the biochemical characteristics of the small laccase from ...
|
|
69742
|
The effect of mutations near the T1 copper site on the biochemical characteristics of the small laccase from ...
|
|
69743
|
Structural and functional characterization of two-domain laccase from Streptomyces viridochromogenes
|
|
69744
|
Structural and functional characterization of two-domain laccase from Streptomyces viridochromogenes
|
|
69745
|
Structural and functional characterization of two-domain laccase from Streptomyces viridochromogenes
|
|
69746
|
Structural and functional characterization of two-domain laccase from Streptomyces viridochromogenes
|
|
69747
|
Structural and functional characterization of two-domain laccase from Streptomyces viridochromogenes
|
|
69748
|
Structural and functional characterization of two-domain laccase from Streptomyces viridochromogenes
|
|
69749
|
Expanding the laccase- toolbox: a laccase from Corynebacterium glutamicum with phenol coupling and cuprous ...
|
|
69750
|
Expanding the laccase- toolbox: a laccase from Corynebacterium glutamicum with phenol coupling and cuprous ...
|
|
69751
|
Expanding the laccase- toolbox: a laccase from Corynebacterium glutamicum with phenol coupling and cuprous ...
|
|
69752
|
Expanding the laccase- toolbox: a laccase from Corynebacterium glutamicum with phenol coupling and cuprous ...
|
|
69753
|
Structure- based rational design to enhance the solubility and thermostability of a bacterial laccase Lac15
|
|
69754
|
Structure- based rational design to enhance the solubility and thermostability of a bacterial laccase Lac15
|
|
69755
|
Structure- based rational design to enhance the solubility and thermostability of a bacterial laccase Lac15
|
|
69756
|
Overexpression of a novel thermostable and chloride-tolerant laccase from Thermus thermophilus SG0.5JP17-16 in ...
|
|
69757
|
Overexpression of a novel thermostable and chloride-tolerant laccase from Thermus thermophilus SG0.5JP17-16 in ...
|
|
69758
|
Overexpression of a novel thermostable and chloride-tolerant laccase from Thermus thermophilus SG0.5JP17-16 in ...
|
|
69759
|
Overexpression of a novel thermostable and chloride-tolerant laccase from Thermus thermophilus SG0.5JP17-16 in ...
|
|
69760
|
Expression of CotA laccase in Pichia pastoris and its electrocatalytic sensing application for hydrogen ...
|
|
69761
|
Expression of CotA laccase in Pichia pastoris and its electrocatalytic sensing application for hydrogen ...
|
|
69762
|
Expression of CotA laccase in Pichia pastoris and its electrocatalytic sensing application for hydrogen ...
|
|
69763
|
Improving the Indigo Carmine Decolorization Ability of a Bacillus amyloliquefaciens Laccase by Site-Directed ...
|
|
69764
|
Improving the Indigo Carmine Decolorization Ability of a Bacillus amyloliquefaciens Laccase by Site-Directed ...
|
|
69765
|
Identification of bacterial laccase cueO mutation from the metagenome of chemical plant sludge
|
|
69766
|
Identification of bacterial laccase cueO mutation from the metagenome of chemical plant sludge
|
|
69767
|
Identification of bacterial laccase cueO mutation from the metagenome of chemical plant sludge
|
|
69768
|
Identification of bacterial laccase cueO mutation from the metagenome of chemical plant sludge
|
|
69769
|
Identification of bacterial laccase cueO mutation from the metagenome of chemical plant sludge
|
|
69770
|
Combined strategies for improving production of a thermo-alkali stable laccase in Pichia pastoris
|
|
69771
|
TtMCO: A highly thermostable laccase-like multicopper oxidase from the thermophilic Thermobaculum terrenum
|
|
69772
|
TtMCO: A highly thermostable laccase-like multicopper oxidase from the thermophilic Thermobaculum terrenum
|
|
69773
|
TtMCO: A highly thermostable laccase-like multicopper oxidase from the thermophilic Thermobaculum terrenum
|
|
69774
|
TtMCO: A highly thermostable laccase-like multicopper oxidase from the thermophilic Thermobaculum terrenum
|
|
69775
|
TtMCO: A highly thermostable laccase-like multicopper oxidase from the thermophilic Thermobaculum terrenum
|
|
69776
|
Characterization of a low redox potential laccase from the basidiomycete C30
|
|
69777
|
Characterization of a low redox potential laccase from the basidiomycete C30
|
|
69778
|
Characterization of a low redox potential laccase from the basidiomycete C30
|
|
69779
|
Characterization of a low redox potential laccase from the basidiomycete C30
|
|
69780
|
Characterization of a low redox potential laccase from the basidiomycete C30
|
|
69781
|
Characterization of a low redox potential laccase from the basidiomycete C30
|
|
69782
|
Characterization of a low redox potential laccase from the basidiomycete C30
|
|
69783
|
Characterization of a low redox potential laccase from the basidiomycete C30
|
|
69784
|
Characterization of a low redox potential laccase from the basidiomycete C30
|
|
69785
|
Characterization of a low redox potential laccase from the basidiomycete C30
|
|
69786
|
Molecular and biochemical characterization of a new thermostable bacterial laccase from Meiothermus ruber DSM ...
|
|
69787
|
Molecular and biochemical characterization of a new thermostable bacterial laccase from Meiothermus ruber DSM ...
|
|
69788
|
Molecular and biochemical characterization of a new thermostable bacterial laccase from Meiothermus ruber DSM ...
|
|
69789
|
Multicopper oxidase mutant with improved salt tolerance
|
|
69790
|
Multicopper oxidase mutant with improved salt tolerance
|
|
69791
|
Multicopper oxidase mutant with improved salt tolerance
|
|
69792
|
Multicopper oxidase mutant with improved salt tolerance
|
|
69793
|
Multicopper oxidase mutant with improved salt tolerance
|
|
69794
|
Partial purification and characterization of the novel halotolerant and alkalophilic laccase produced by a new ...
|
|
69795
|
Production Characteristics, Activity Patterns and Biodecolourisation Applications of Thermostable Laccases ...
|
|
69796
|
Production Characteristics, Activity Patterns and Biodecolourisation Applications of Thermostable Laccases ...
|
|
69797
|
Enhanced expression of a recombinant multicopper oxidase, CueO, from Escherichia coli and its laccase activity ...
|
|
69798
|
Enhanced expression of a recombinant multicopper oxidase, CueO, from Escherichia coli and its laccase activity ...
|
|
69799
|
Enhanced expression of a recombinant multicopper oxidase, CueO, from Escherichia coli and its laccase activity ...
|
|
69800
|
Enhanced expression of a recombinant multicopper oxidase, CueO, from Escherichia coli and its laccase activity ...
|
|
69801
|
Enhanced expression of a recombinant multicopper oxidase, CueO, from Escherichia coli and its laccase activity ...
|
|
69802
|
Enhanced expression of a recombinant multicopper oxidase, CueO, from Escherichia coli and its laccase activity ...
|
|
69803
|
High-level expression of Bacillus amyloliquefaciens laccase and construction of its chimeric variant with ...
|
|
69804
|
Characterization and application of a novel laccase derived from Bacillus amyloliquefaciens
|
|
69805
|
Dual Role of Multicopper Oxidase of Klebsiella pneumoniae as a Copper Homeostatic Element and a Novel Alkaline ...
|
|
69806
|
Dual Role of Multicopper Oxidase of Klebsiella pneumoniae as a Copper Homeostatic Element and a Novel Alkaline ...
|
|
69807
|
Exploring the lignolytic potential of a new laccase producing strain Kocuria sp PBS-1 and its application in ...
|
|
69808
|
Deletion and site-directed mutagenesis of laccase from Shigella dysenteriae results in enhanced enzymatic ...
|
|
69809
|
Deletion and site-directed mutagenesis of laccase from Shigella dysenteriae results in enhanced enzymatic ...
|
|
69810
|
Deletion and site-directed mutagenesis of laccase from Shigella dysenteriae results in enhanced enzymatic ...
|
|
69811
|
A non-blue laccase of Bacillus sp. GZB displays manganese-oxidase activity: A study of laccase ...
|
|
69812
|
A non-blue laccase of Bacillus sp. GZB displays manganese-oxidase activity: A study of laccase ...
|
|
69813
|
A non-blue laccase of Bacillus sp. GZB displays manganese-oxidase activity: A study of laccase ...
|
|
69814
|
Transformation of humic acids by two-domain laccase from Streptomyces anulatus
|
|
69815
|
Transformation of humic acids by two-domain laccase from Streptomyces anulatus
|
|
69816
|
Transformation of humic acids by two-domain laccase from Streptomyces anulatus
|
|
69817
|
Transformation of humic acids by two-domain laccase from Streptomyces anulatus
|
|
69818
|
Insight into Depolymerization Mechanism of Bacterial Laccase for Lignin
|
|
69819
|
Insight into Depolymerization Mechanism of Bacterial Laccase for Lignin
|
|
69820
|
Insight into Depolymerization Mechanism of Bacterial Laccase for Lignin
|
|
69821
|
Insight into Depolymerization Mechanism of Bacterial Laccase for Lignin
|
|
69822
|
Extracellular Thermostable Laccase-Like Enzymes from Bacillus licheniformis Strains: Production, Purification ...
|
|
69823
|
Extracellular Thermostable Laccase-Like Enzymes from Bacillus licheniformis Strains: Production, Purification ...
|
|
69824
|
Extracellular Thermostable Laccase-Like Enzymes from Bacillus licheniformis Strains: Production, Purification ...
|
|
69825
|
Extracellular Thermostable Laccase-Like Enzymes from Bacillus licheniformis Strains: Production, Purification ...
|
|
69826
|
Catalytic oxidation of manganese(II) by multicopper oxidase CueO and characterization of the biogenic Mn oxide
|
|
69827
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69828
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69829
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69830
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69831
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69832
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69833
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69834
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69835
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69836
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69837
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69838
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69839
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69840
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69841
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69842
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69843
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69844
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69845
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69846
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69847
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69848
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69849
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69850
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69851
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69852
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69853
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69854
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69855
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69856
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69857
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69858
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69859
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69860
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69861
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69862
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69863
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69864
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69865
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69866
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69867
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69868
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69869
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69870
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69871
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69872
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69873
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69874
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69875
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69876
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69877
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69878
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69879
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69880
|
Turning a Hyperthermostable Metallo-Oxidase into a Laccase by Directed Evolution
|
|
69881
|
Cloning and characterization of a new laccase from Bacillus licheniformis catalyzing dimerization of phenolic ...
|
|
69882
|
Cloning and characterization of a new laccase from Bacillus licheniformis catalyzing dimerization of phenolic ...
|
|
69883
|
Cloning and characterization of a new laccase from Bacillus licheniformis catalyzing dimerization of phenolic ...
|
|
69884
|
Improving the functional expression of a Bacillus licheniformis laccase by random and site-directed ...
|
|
69885
|
Improving the functional expression of a Bacillus licheniformis laccase by random and site-directed ...
|
|
69886
|
Improving the functional expression of a Bacillus licheniformis laccase by random and site-directed ...
|
|
69887
|
Improving the functional expression of a Bacillus licheniformis laccase by random and site-directed ...
|
|
69888
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69889
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69890
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69891
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69892
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69893
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69894
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69895
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69896
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69897
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69898
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69899
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69900
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69901
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69902
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69903
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69904
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69905
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69906
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69907
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69908
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69909
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69910
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69911
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69912
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69913
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69914
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69915
|
Protein engineering of laccase to enhance its activity and stability in the presence of organic solvents
|
|
69916
|
Correlation between the T1 copper reduction potential and catalytic activity of a small laccase
|
|
69917
|
Correlation between the T1 copper reduction potential and catalytic activity of a small laccase
|
|
69918
|
Correlation between the T1 copper reduction potential and catalytic activity of a small laccase
|
|
69919
|
Correlation between the T1 copper reduction potential and catalytic activity of a small laccase
|
|
69920
|
Correlation between the T1 copper reduction potential and catalytic activity of a small laccase
|
|
69921
|
Correlation between the T1 copper reduction potential and catalytic activity of a small laccase
|
|
69922
|
Correlation between the T1 copper reduction potential and catalytic activity of a small laccase
|
|
69923
|
Correlation between the T1 copper reduction potential and catalytic activity of a small laccase
|
|
69924
|
Correlation between the T1 copper reduction potential and catalytic activity of a small laccase
|
|
69925
|
Correlation between the T1 copper reduction potential and catalytic activity of a small laccase
|
|
69926
|
Correlation between the T1 copper reduction potential and catalytic activity of a small laccase
|
|
69927
|
Correlation between the T1 copper reduction potential and catalytic activity of a small laccase
|
|
69928
|
Correlation between the T1 copper reduction potential and catalytic activity of a small laccase
|
|
69929
|
Correlation between the T1 copper reduction potential and catalytic activity of a small laccase
|
|
69930
|
Cloning of multicopper oxidase gene from Ochrobactrum sp. 531 and characterization of its alkaline laccase ...
|
|
69931
|
Cloning of multicopper oxidase gene from Ochrobactrum sp. 531 and characterization of its alkaline laccase ...
|
|
69932
|
Cloning of multicopper oxidase gene from Ochrobactrum sp. 531 and characterization of its alkaline laccase ...
|
|
69933
|
Electrochemical properties and temperature dependence of a recombinant laccase from Thermus thermophilus
|
|
69934
|
Electrochemical properties and temperature dependence of a recombinant laccase from Thermus thermophilus
|
|
69935
|
Electrochemical properties and temperature dependence of a recombinant laccase from Thermus thermophilus
|
|
69936
|
Electrochemical properties and temperature dependence of a recombinant laccase from Thermus thermophilus
|
|
69937
|
Electrochemical properties and temperature dependence of a recombinant laccase from Thermus thermophilus
|
|
69938
|
Electrochemical properties and temperature dependence of a recombinant laccase from Thermus thermophilus
|
|
69939
|
Enhancement of laccase activity through the construction and breakdown of a hydrogen bond at the type I copper ...
|
|
69940
|
Enhancement of laccase activity through the construction and breakdown of a hydrogen bond at the type I copper ...
|
|
69941
|
Enhancement of laccase activity through the construction and breakdown of a hydrogen bond at the type I copper ...
|
|
69942
|
Enhancement of laccase activity through the construction and breakdown of a hydrogen bond at the type I copper ...
|
|
69943
|
Enhancement of laccase activity through the construction and breakdown of a hydrogen bond at the type I copper ...
|
|
69944
|
Enhancement of laccase activity through the construction and breakdown of a hydrogen bond at the type I copper ...
|
|
69945
|
Enhancement of laccase activity through the construction and breakdown of a hydrogen bond at the type I copper ...
|
|
69946
|
Enhancement of laccase activity through the construction and breakdown of a hydrogen bond at the type I copper ...
|
|
69947
|
Enhancement of laccase activity through the construction and breakdown of a hydrogen bond at the type I copper ...
|
|
69948
|
Enhancement of laccase activity through the construction and breakdown of a hydrogen bond at the type I copper ...
|
|
69949
|
Enhancement of laccase activity through the construction and breakdown of a hydrogen bond at the type I copper ...
|
|
69950
|
Enhancement of laccase activity through the construction and breakdown of a hydrogen bond at the type I copper ...
|
|
69951
|
Characterization of a laccase-like multicopper oxidase from newly isolated Streptomyces sp. C1 in agricultural ...
|
|
69952
|
Characterization of a laccase-like multicopper oxidase from newly isolated Streptomyces sp. C1 in agricultural ...
|
|
69953
|
Characterization of a laccase-like multicopper oxidase from newly isolated Streptomyces sp. C1 in agricultural ...
|
|
69954
|
Characterization of a laccase-like multicopper oxidase from newly isolated Streptomyces sp. C1 in agricultural ...
|
|
69955
|
Cell thermolysis - A simple and fast approach for isolation of bacterial laccases with potential to decolorize ...
|
|
69956
|
Cell thermolysis - A simple and fast approach for isolation of bacterial laccases with potential to decolorize ...
|
|
69957
|
Cell thermolysis - A simple and fast approach for isolation of bacterial laccases with potential to decolorize ...
|
|
69958
|
Cell thermolysis - A simple and fast approach for isolation of bacterial laccases with potential to decolorize ...
|
|
69959
|
Cell thermolysis - A simple and fast approach for isolation of bacterial laccases with potential to decolorize ...
|
|
69960
|
Cell thermolysis - A simple and fast approach for isolation of bacterial laccases with potential to decolorize ...
|
|
69961
|
Structural and functional characterisation of multi-copper oxidase CueO from lignin-degrading bacterium ...
|
|
69962
|
Structural and functional characterisation of multi-copper oxidase CueO from lignin-degrading bacterium ...
|
|
69963
|
Structural and functional characterisation of multi-copper oxidase CueO from lignin-degrading bacterium ...
|
|
69964
|
An enzymatic system for decolorization of wastewater dyes using immobilized CueO laccase-like multicopper ...
|
|
69965
|
An enzymatic system for decolorization of wastewater dyes using immobilized CueO laccase-like multicopper ...
|
|
69966
|
An enzymatic system for decolorization of wastewater dyes using immobilized CueO laccase-like multicopper ...
|
|
69967
|
Characterization of multicopper oxidase CopA from Pseudomonas putida KT2440 and Pseudomonas fluorescens Pf-5: ...
|
|
69968
|
Characterization of multicopper oxidase CopA from Pseudomonas putida KT2440 and Pseudomonas fluorescens Pf-5: ...
|
|
69969
|
Characterization of multicopper oxidase CopA from Pseudomonas putida KT2440 and Pseudomonas fluorescens Pf-5: ...
|
|
69970
|
Characterization of multicopper oxidase CopA from Pseudomonas putida KT2440 and Pseudomonas fluorescens Pf-5: ...
|
|
69971
|
Characterization of multicopper oxidase CopA from Pseudomonas putida KT2440 and Pseudomonas fluorescens Pf-5: ...
|
|
69972
|
Characterization of multicopper oxidase CopA from Pseudomonas putida KT2440 and Pseudomonas fluorescens Pf-5: ...
|
|
69973
|
Transcriptional analysis of the laccase-like gene from Burkholderia cepacia BNS and expression in Escherichia ...
|
|
69974
|
Transcriptional analysis of the laccase-like gene from Burkholderia cepacia BNS and expression in Escherichia ...
|
|
69975
|
Functional substitution of domain 3 (T1 copper center) of a novel laccase with Cu ions
|
|
69976
|
Functional substitution of domain 3 (T1 copper center) of a novel laccase with Cu ions
|
|
69977
|
Functional substitution of domain 3 (T1 copper center) of a novel laccase with Cu ions
|
|
69978
|
Functional substitution of domain 3 (T1 copper center) of a novel laccase with Cu ions
|
|
69979
|
Functional substitution of domain 3 (T1 copper center) of a novel laccase with Cu ions
|
|
69980
|
Functional substitution of domain 3 (T1 copper center) of a novel laccase with Cu ions
|
|
69981
|
Functional substitution of domain 3 (T1 copper center) of a novel laccase with Cu ions
|
|
69982
|
Functional substitution of domain 3 (T1 copper center) of a novel laccase with Cu ions
|
|
69983
|
Functional substitution of domain 3 (T1 copper center) of a novel laccase with Cu ions
|
|
69984
|
Functional substitution of domain 3 (T1 copper center) of a novel laccase with Cu ions
|
|
69985
|
An extracellular thermo-alkali-stable laccase from Bacillus tequilensis SN4, with a potential to biobleach ...
|
|
69986
|
An extracellular thermo-alkali-stable laccase from Bacillus tequilensis SN4, with a potential to biobleach ...
|
|
69987
|
Biochemical characterization of an extremely stable pH-versatile laccase from Sporothrix carnis CPF-05.
|
|
69988
|
Preparation and characterization of stable cross-linked enzyme aggregates of novel laccase enzyme from ...
|
|
69989
|
Preparation and characterization of stable cross-linked enzyme aggregates of novel laccase enzyme from ...
|
|
69990
|
Refolding, characterization, and dye decolorization ability of a highly thermostable laccase from Geobacillus ...
|
|
69991
|
Refolding, characterization, and dye decolorization ability of a highly thermostable laccase from Geobacillus ...
|
|
69992
|
Refolding, characterization, and dye decolorization ability of a highly thermostable laccase from Geobacillus ...
|
|
69993
|
Oxygen- reducing enzyme cathodes produced from SLAC, a small laccase from Streptomyces coelicolor
|
|
69994
|
Oxygen- reducing enzyme cathodes produced from SLAC, a small laccase from Streptomyces coelicolor
|
|
69995
|
Characterization of SLAC: a small laccase from Streptomyces coelicolor with unprecedented activity
|
|
69996
|
Iodide oxidation by a novel multicopper oxidase from the alphaproteobacterium strain Q-1.
|
|
69997
|
Iodide oxidation by a novel multicopper oxidase from the alphaproteobacterium strain Q-1.
|
|
69998
|
Multicopper oxidase of Acinetobacter baumannii: Assessing its role in metal homeostasis, stress management and ...
|
|
69999
|
Multicopper oxidase of Acinetobacter baumannii: Assessing its role in metal homeostasis, stress management and ...
|
|
70000
|
Multicopper oxidase of Acinetobacter baumannii: Assessing its role in metal homeostasis, stress management and ...
|





 If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.
If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.