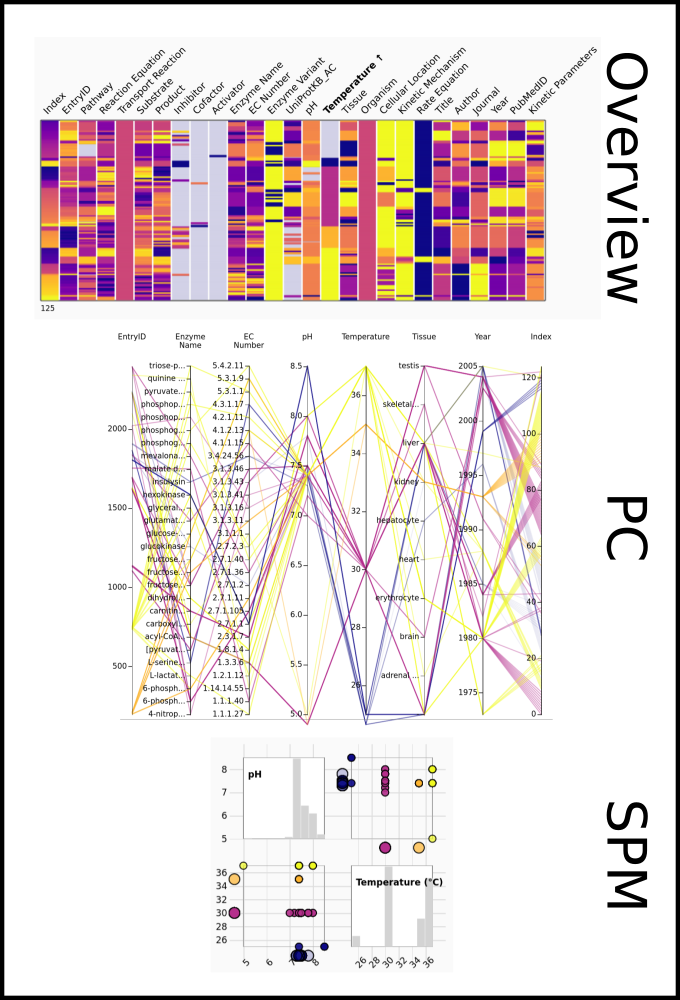
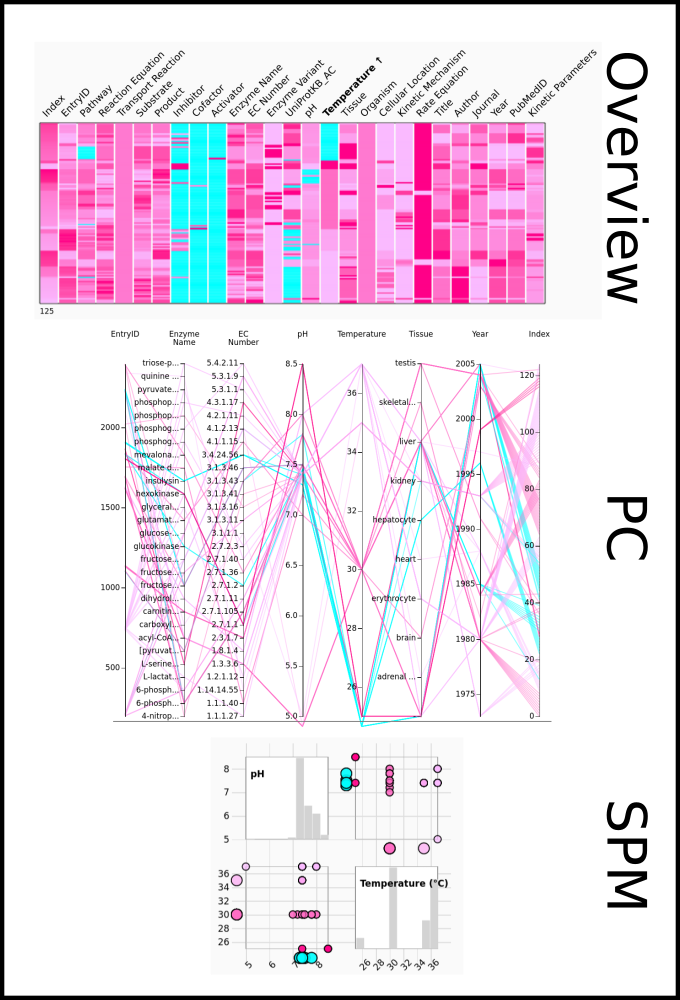
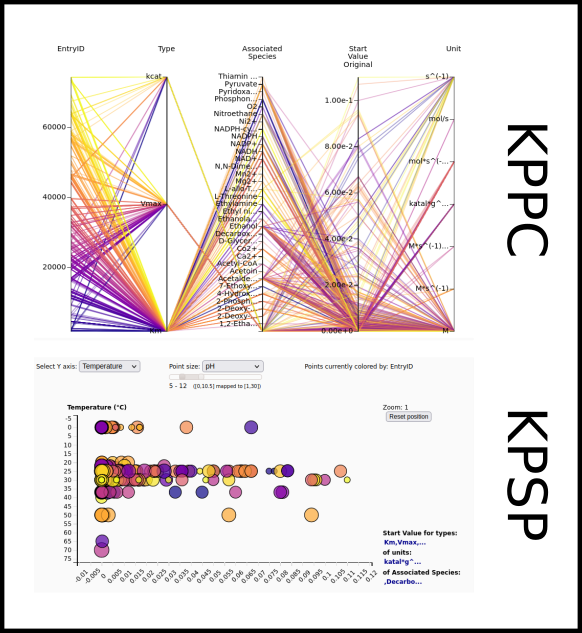
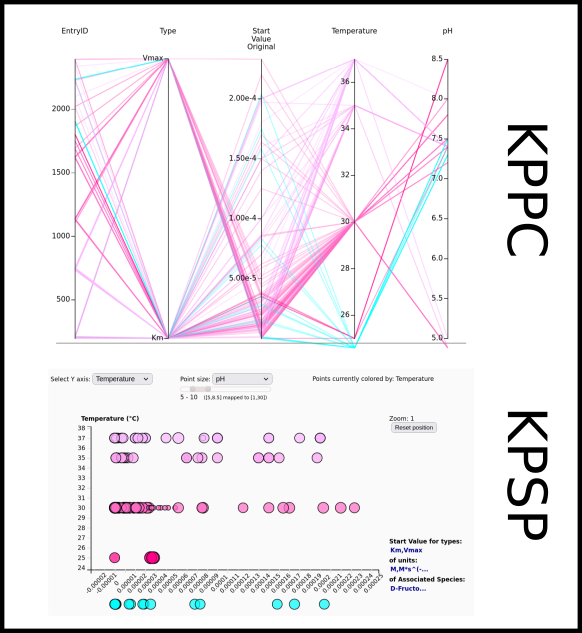
To improve SABIO-RK data interoperability, semantic markup was added to web pages as described and defined by
This structured information makes it easier to discover, collate and analyse our data.
|
68001
|
PPi-dependent phosphofructokinase from Thermoproteus tenax, an archaeal descendant of an ancient line in ...
|
|
68002
|
Enzymatic characteristics of recombinant medium isozyme of 2'-5' oligoadenylate synthetase.
|
|
68003
|
A chloroplastic UDP-glucose pyrophosphorylase from Arabidopsis is the committed enzyme for the first step of ...
|
|
68004
|
A chloroplastic UDP-glucose pyrophosphorylase from Arabidopsis is the committed enzyme for the first step of ...
|
|
68005
|
Functional comparison of citrate synthase isoforms from S. cerevisiae.
|
|
68006
|
Functional comparison of citrate synthase isoforms from S. cerevisiae.
|
|
68007
|
Functional comparison of citrate synthase isoforms from S. cerevisiae.
|
|
68008
|
Deletion mutagenesis using an 'M13 splint': the N-terminal structural domain of tyrosyl-tRNA synthetase (B. ...
|
|
68009
|
Deletion mutagenesis using an 'M13 splint': the N-terminal structural domain of tyrosyl-tRNA synthetase (B. ...
|
|
68010
|
Deletion mutagenesis using an 'M13 splint': the N-terminal structural domain of tyrosyl-tRNA synthetase (B. ...
|
|
68011
|
Deletion mutagenesis using an 'M13 splint': the N-terminal structural domain of tyrosyl-tRNA synthetase (B. ...
|
|
68012
|
Mechanism of molecular interactions for tRNA-Val recognition by valyl-tRNA synthetase
|
|
68013
|
Mechanism of molecular interactions for tRNA-Val recognition by valyl-tRNA synthetase
|
|
68014
|
Mechanism of molecular interactions for tRNA-Val recognition by valyl-tRNA synthetase
|
|
68015
|
Mechanism of molecular interactions for tRNA-Val recognition by valyl-tRNA synthetase
|
|
68016
|
Mechanism of molecular interactions for tRNA-Val recognition by valyl-tRNA synthetase
|
|
68017
|
Mechanism of molecular interactions for tRNA-Val recognition by valyl-tRNA synthetase
|
|
68018
|
Mechanism of molecular interactions for tRNA-Val recognition by valyl-tRNA synthetase
|
|
68019
|
Mechanism of molecular interactions for tRNA-Val recognition by valyl-tRNA synthetase
|
|
68020
|
Mechanism of molecular interactions for tRNA-Val recognition by valyl-tRNA synthetase
|
|
68021
|
Mechanism of molecular interactions for tRNA-Val recognition by valyl-tRNA synthetase
|
|
68022
|
Re-citrate synthase from Clostridium kluyveri is phylogenetically related to homocitrate synthase and ...
|
|
68023
|
Re-citrate synthase from Clostridium kluyveri is phylogenetically related to homocitrate synthase and ...
|
|
68024
|
Re-citrate synthase from Clostridium kluyveri is phylogenetically related to homocitrate synthase and ...
|
|
68025
|
Rat malonyl-CoA decarboxylase; cloning, expression in E. coli and its biochemical characterization.
|
|
68026
|
Cocrystal structures of diaminopimelate decarboxylase: mechanism, evolution, and inhibition of an antibiotic ...
|
|
68027
|
Cocrystal structures of diaminopimelate decarboxylase: mechanism, evolution, and inhibition of an antibiotic ...
|
|
68028
|
Cocrystal structures of diaminopimelate decarboxylase: mechanism, evolution, and inhibition of an antibiotic ...
|
|
68029
|
Cocrystal structures of diaminopimelate decarboxylase: mechanism, evolution, and inhibition of an antibiotic ...
|
|
68030
|
Cocrystal structures of diaminopimelate decarboxylase: mechanism, evolution, and inhibition of an antibiotic ...
|
|
68031
|
Cocrystal structures of diaminopimelate decarboxylase: mechanism, evolution, and inhibition of an antibiotic ...
|
|
68032
|
Cocrystal structures of diaminopimelate decarboxylase: mechanism, evolution, and inhibition of an antibiotic ...
|
|
68033
|
Cocrystal structures of diaminopimelate decarboxylase: mechanism, evolution, and inhibition of an antibiotic ...
|
|
68034
|
Cocrystal structures of diaminopimelate decarboxylase: mechanism, evolution, and inhibition of an antibiotic ...
|
|
68035
|
Plants synthesize ethanolamine by direct decarboxylation of serine using a pyridoxal phosphate enzyme.
|
|
68036
|
Plants synthesize ethanolamine by direct decarboxylation of serine using a pyridoxal phosphate enzyme.
|
|
68037
|
Plants synthesize ethanolamine by direct decarboxylation of serine using a pyridoxal phosphate enzyme.
|
|
68038
|
Partial Purification and Characterization of Red Chlorophyll Catabolite Reductase, a Stroma Protein Involved ...
|
|
68039
|
Allosteric control by calcium and mechanism of desensitization of phosphoenolpyruvate carboxykinase of ...
|
|
68040
|
Allosteric control by calcium and mechanism of desensitization of phosphoenolpyruvate carboxykinase of ...
|
|
68041
|
Allosteric control by calcium and mechanism of desensitization of phosphoenolpyruvate carboxykinase of ...
|
|
68042
|
Allosteric control by calcium and mechanism of desensitization of phosphoenolpyruvate carboxykinase of ...
|
|
68043
|
Allosteric control by calcium and mechanism of desensitization of phosphoenolpyruvate carboxykinase of ...
|
|
68044
|
Allosteric control by calcium and mechanism of desensitization of phosphoenolpyruvate carboxykinase of ...
|
|
68045
|
Allosteric control by calcium and mechanism of desensitization of phosphoenolpyruvate carboxykinase of ...
|
|
68046
|
Allosteric control by calcium and mechanism of desensitization of phosphoenolpyruvate carboxykinase of ...
|
|
68047
|
Allosteric control by calcium and mechanism of desensitization of phosphoenolpyruvate carboxykinase of ...
|
|
68048
|
Allosteric control by calcium and mechanism of desensitization of phosphoenolpyruvate carboxykinase of ...
|
|
68049
|
Allosteric control by calcium and mechanism of desensitization of phosphoenolpyruvate carboxykinase of ...
|
|
68050
|
Allosteric control by calcium and mechanism of desensitization of phosphoenolpyruvate carboxykinase of ...
|
|
68051
|
Allosteric control by calcium and mechanism of desensitization of phosphoenolpyruvate carboxykinase of ...
|
|
68052
|
Allosteric control by calcium and mechanism of desensitization of phosphoenolpyruvate carboxykinase of ...
|
|
68053
|
Allosteric control by calcium and mechanism of desensitization of phosphoenolpyruvate carboxykinase of ...
|
|
68054
|
Allosteric control by calcium and mechanism of desensitization of phosphoenolpyruvate carboxykinase of ...
|
|
68055
|
Allosteric control by calcium and mechanism of desensitization of phosphoenolpyruvate carboxykinase of ...
|
|
68056
|
EH3 (ABHD9): the first member of a new epoxide hydrolase family with high activity for fatty acid epoxides.
|
|
68057
|
EH3 (ABHD9): the first member of a new epoxide hydrolase family with high activity for fatty acid epoxides.
|
|
68058
|
EH3 (ABHD9): the first member of a new epoxide hydrolase family with high activity for fatty acid epoxides.
|
|
68059
|
EH3 (ABHD9): the first member of a new epoxide hydrolase family with high activity for fatty acid epoxides.
|
|
68060
|
EH3 (ABHD9): the first member of a new epoxide hydrolase family with high activity for fatty acid epoxides.
|
|
68061
|
EH3 (ABHD9): the first member of a new epoxide hydrolase family with high activity for fatty acid epoxides.
|
|
68062
|
EH3 (ABHD9): the first member of a new epoxide hydrolase family with high activity for fatty acid epoxides.
|
|
68063
|
EH3 (ABHD9): the first member of a new epoxide hydrolase family with high activity for fatty acid epoxides.
|
|
68064
|
EH3 (ABHD9): the first member of a new epoxide hydrolase family with high activity for fatty acid epoxides.
|
|
68065
|
EH3 (ABHD9): the first member of a new epoxide hydrolase family with high activity for fatty acid epoxides.
|
|
68066
|
EH3 (ABHD9): the first member of a new epoxide hydrolase family with high activity for fatty acid epoxides.
|
|
68067
|
EH3 (ABHD9): the first member of a new epoxide hydrolase family with high activity for fatty acid epoxides.
|
|
68068
|
EH3 (ABHD9): the first member of a new epoxide hydrolase family with high activity for fatty acid epoxides.
|
|
68069
|
EH3 (ABHD9): the first member of a new epoxide hydrolase family with high activity for fatty acid epoxides.
|
|
68070
|
EH3 (ABHD9): the first member of a new epoxide hydrolase family with high activity for fatty acid epoxides.
|
|
68071
|
A methylaspartate cycle in haloarchaea.
|
|
68072
|
A methylaspartate cycle in haloarchaea.
|
|
68073
|
A methylaspartate cycle in haloarchaea.
|
|
68074
|
A methylaspartate cycle in haloarchaea.
|
|
68075
|
A methylaspartate cycle in haloarchaea.
|
|
68076
|
A methylaspartate cycle in haloarchaea.
|
|
68077
|
A methylaspartate cycle in haloarchaea.
|
|
68078
|
A methylaspartate cycle in haloarchaea.
|
|
68079
|
A methylaspartate cycle in haloarchaea.
|
|
68080
|
A methylaspartate cycle in haloarchaea.
|
|
68081
|
A methylaspartate cycle in haloarchaea.
|
|
68082
|
A methylaspartate cycle in haloarchaea.
|
|
68083
|
A methylaspartate cycle in haloarchaea.
|
|
68084
|
A genetic locus required for iron acquisition in Mycobacterium tuberculosis
|
|
68085
|
A genetic locus required for iron acquisition in Mycobacterium tuberculosis
|
|
68086
|
A genetic locus required for iron acquisition in Mycobacterium tuberculosis
|
|
68087
|
A genetic locus required for iron acquisition in Mycobacterium tuberculosis
|
|
68088
|
A genetic locus required for iron acquisition in Mycobacterium tuberculosis
|
|
68089
|
PapA1 and PapA2 are acyltransferases essential for the biosynthesis of the Mycobacterium tuberculosis ...
|
|
68090
|
PapA1 and PapA2 are acyltransferases essential for the biosynthesis of the Mycobacterium tuberculosis ...
|
|
68091
|
PapA1 and PapA2 are acyltransferases essential for the biosynthesis of the Mycobacterium tuberculosis ...
|
|
68092
|
Purification and properties of NADP(+)-dependent isocitrate dehydrogenase from the corpus luteum
|
|
68093
|
Purification and physicochemical characterization of chicken heart citrate synthase.
|
|
68094
|
Escherichia coli citrate synthase. Purification and the effect of potassium on some properties.
|
|
68095
|
Escherichia coli citrate synthase. Purification and the effect of potassium on some properties.
|
|
68096
|
Kinetic studies on citrate synthase from pig heart.
|
|
68097
|
Studies on two isozymes of aconitase from Bacillus cereus T. III. Enzymatic properties.
|
|
68098
|
Studies on two isozymes of aconitase from Bacillus cereus T. III. Enzymatic properties.
|
|
68099
|
Studies on two isozymes of aconitase from Bacillus cereus T. III. Enzymatic properties.
|
|
68100
|
Studies on two isozymes of aconitase from Bacillus cereus T. III. Enzymatic properties.
|
|
68101
|
Evidence for the identity and some comparative properties of alpha-ketoglutarate and 2-keto-4-hydroxyglutarate ...
|
|
68102
|
Evidence for the identity and some comparative properties of alpha-ketoglutarate and 2-keto-4-hydroxyglutarate ...
|
|
68103
|
Evidence for the identity and some comparative properties of alpha-ketoglutarate and 2-keto-4-hydroxyglutarate ...
|
|
68104
|
Evidence for the identity and some comparative properties of alpha-ketoglutarate and 2-keto-4-hydroxyglutarate ...
|
|
68105
|
Evidence for the identity and some comparative properties of alpha-ketoglutarate and 2-keto-4-hydroxyglutarate ...
|
|
68106
|
Evidence for the identity and some comparative properties of alpha-ketoglutarate and 2-keto-4-hydroxyglutarate ...
|
|
68107
|
Evidence for the identity and some comparative properties of alpha-ketoglutarate and 2-keto-4-hydroxyglutarate ...
|
|
68108
|
Evidence for the identity and some comparative properties of alpha-ketoglutarate and 2-keto-4-hydroxyglutarate ...
|
|
68109
|
Purification and properties of NADP+:isocitrate dehydrogenase from lactating bovine mammary gland.
|
|
68110
|
Purification and properties of NADP+:isocitrate dehydrogenase from lactating bovine mammary gland.
|
|
68111
|
Purification and properties of NADP+:isocitrate dehydrogenase from lactating bovine mammary gland.
|
|
68112
|
Purification and properties of NADP+:isocitrate dehydrogenase from lactating bovine mammary gland.
|
|
68113
|
Purification and properties of NADP+:isocitrate dehydrogenase from lactating bovine mammary gland.
|
|
68114
|
Biochemical characterization of dTDP-D-Qui4N and dTDP-D-Qui4NAc biosynthetic pathways in Shigella dysenteriae ...
|
|
68115
|
Biochemical characterization of dTDP-D-Qui4N and dTDP-D-Qui4NAc biosynthetic pathways in Shigella dysenteriae ...
|
|
68116
|
Biochemical characterization of dTDP-D-Qui4N and dTDP-D-Qui4NAc biosynthetic pathways in Shigella dysenteriae ...
|
|
68117
|
Biochemical characterization of dTDP-D-Qui4N and dTDP-D-Qui4NAc biosynthetic pathways in Shigella dysenteriae ...
|
|
68118
|
Inhibition of the Escherichia coli pyruvate dehydrogenase complex E1 subunit and its tyrosine 177 variants by ...
|
|
68119
|
Inhibition of the Escherichia coli pyruvate dehydrogenase complex E1 subunit and its tyrosine 177 variants by ...
|
|
68120
|
Inhibition of the Escherichia coli pyruvate dehydrogenase complex E1 subunit and its tyrosine 177 variants by ...
|
|
68121
|
Inhibition of the Escherichia coli pyruvate dehydrogenase complex E1 subunit and its tyrosine 177 variants by ...
|
|
68122
|
Inhibition of the Escherichia coli pyruvate dehydrogenase complex E1 subunit and its tyrosine 177 variants by ...
|
|
68123
|
Inhibition of the Escherichia coli pyruvate dehydrogenase complex E1 subunit and its tyrosine 177 variants by ...
|
|
68124
|
Inhibition of the Escherichia coli pyruvate dehydrogenase complex E1 subunit and its tyrosine 177 variants by ...
|
|
68125
|
Inhibition of the Escherichia coli pyruvate dehydrogenase complex E1 subunit and its tyrosine 177 variants by ...
|
|
68126
|
Inhibition of the Escherichia coli pyruvate dehydrogenase complex E1 subunit and its tyrosine 177 variants by ...
|
|
68127
|
Inhibition of the Escherichia coli pyruvate dehydrogenase complex E1 subunit and its tyrosine 177 variants by ...
|
|
68128
|
Inhibition of the Escherichia coli pyruvate dehydrogenase complex E1 subunit and its tyrosine 177 variants by ...
|
|
68129
|
Inhibition of the Escherichia coli pyruvate dehydrogenase complex E1 subunit and its tyrosine 177 variants by ...
|
|
68130
|
Inhibition of the Escherichia coli pyruvate dehydrogenase complex E1 subunit and its tyrosine 177 variants by ...
|
|
68131
|
Inhibition of the Escherichia coli pyruvate dehydrogenase complex E1 subunit and its tyrosine 177 variants by ...
|
|
68132
|
Inhibition of the Escherichia coli pyruvate dehydrogenase complex E1 subunit and its tyrosine 177 variants by ...
|
|
68133
|
Inhibition of the Escherichia coli pyruvate dehydrogenase complex E1 subunit and its tyrosine 177 variants by ...
|
|
68134
|
Inhibition of the Escherichia coli pyruvate dehydrogenase complex E1 subunit and its tyrosine 177 variants by ...
|
|
68135
|
Inhibition of the Escherichia coli pyruvate dehydrogenase complex E1 subunit and its tyrosine 177 variants by ...
|
|
68136
|
Inhibition of the Escherichia coli pyruvate dehydrogenase complex E1 subunit and its tyrosine 177 variants by ...
|
|
68137
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68138
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68139
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68140
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68141
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68142
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68143
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68144
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68145
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68146
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68147
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68148
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68149
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68150
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68151
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68152
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68153
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68154
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68155
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68156
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68157
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68158
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68159
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68160
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68161
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68162
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68163
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68164
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68165
|
Cosubstrate and allosteric modifier activities of structural analogues of NAD and ADP for NAD-specific ...
|
|
68166
|
ACTIVATION AND INHIBITION OF DPN-LINKED ISOCITRATE DEHYDROGENASE OF HEART BY CERTAIN NUCLEOTIDES.
|
|
68167
|
ACTIVATION AND INHIBITION OF DPN-LINKED ISOCITRATE DEHYDROGENASE OF HEART BY CERTAIN NUCLEOTIDES.
|
|
68168
|
ACTIVATION AND INHIBITION OF DPN-LINKED ISOCITRATE DEHYDROGENASE OF HEART BY CERTAIN NUCLEOTIDES.
|
|
68169
|
ACTIVATION AND INHIBITION OF DPN-LINKED ISOCITRATE DEHYDROGENASE OF HEART BY CERTAIN NUCLEOTIDES.
|
|
68170
|
ACTIVATION AND INHIBITION OF DPN-LINKED ISOCITRATE DEHYDROGENASE OF HEART BY CERTAIN NUCLEOTIDES.
|
|
68171
|
ACTIVATION AND INHIBITION OF DPN-LINKED ISOCITRATE DEHYDROGENASE OF HEART BY CERTAIN NUCLEOTIDES.
|
|
68172
|
ACTIVATION AND INHIBITION OF DPN-LINKED ISOCITRATE DEHYDROGENASE OF HEART BY CERTAIN NUCLEOTIDES.
|
|
68173
|
ACTIVATION AND INHIBITION OF DPN-LINKED ISOCITRATE DEHYDROGENASE OF HEART BY CERTAIN NUCLEOTIDES.
|
|
68174
|
ACTIVATION AND INHIBITION OF DPN-LINKED ISOCITRATE DEHYDROGENASE OF HEART BY CERTAIN NUCLEOTIDES.
|
|
68175
|
ACTIVATION AND INHIBITION OF DPN-LINKED ISOCITRATE DEHYDROGENASE OF HEART BY CERTAIN NUCLEOTIDES.
|
|
68176
|
ACTIVATION AND INHIBITION OF DPN-LINKED ISOCITRATE DEHYDROGENASE OF HEART BY CERTAIN NUCLEOTIDES.
|
|
68177
|
ACTIVATION AND INHIBITION OF DPN-LINKED ISOCITRATE DEHYDROGENASE OF HEART BY CERTAIN NUCLEOTIDES.
|
|
68178
|
ACTIVATION AND INHIBITION OF DPN-LINKED ISOCITRATE DEHYDROGENASE OF HEART BY CERTAIN NUCLEOTIDES.
|
|
68179
|
ACTIVATION AND INHIBITION OF DPN-LINKED ISOCITRATE DEHYDROGENASE OF HEART BY CERTAIN NUCLEOTIDES.
|
|
68180
|
ACTIVATION AND INHIBITION OF DPN-LINKED ISOCITRATE DEHYDROGENASE OF HEART BY CERTAIN NUCLEOTIDES.
|
|
68181
|
ACTIVATION AND INHIBITION OF DPN-LINKED ISOCITRATE DEHYDROGENASE OF HEART BY CERTAIN NUCLEOTIDES.
|
|
68182
|
Structure of human dCK suggests strategies to improve anticancer and antiviral therapy
|
|
68183
|
Structure of human dCK suggests strategies to improve anticancer and antiviral therapy
|
|
68184
|
Structure of human dCK suggests strategies to improve anticancer and antiviral therapy
|
|
68185
|
Structure of human dCK suggests strategies to improve anticancer and antiviral therapy
|
|
68186
|
Structure of human dCK suggests strategies to improve anticancer and antiviral therapy
|
|
68187
|
Structure of human dCK suggests strategies to improve anticancer and antiviral therapy
|
|
68188
|
Structure of human dCK suggests strategies to improve anticancer and antiviral therapy
|
|
68189
|
Structure of human dCK suggests strategies to improve anticancer and antiviral therapy
|
|
68190
|
Characterization of two kinases involved in thiamine pyrophosphate and pyridoxal phosphate biosynthesis in ...
|
|
68191
|
Characterization of two kinases involved in thiamine pyrophosphate and pyridoxal phosphate biosynthesis in ...
|
|
68192
|
Isolation and characterization of two fructokinase cDNA clones from rice
|
|
68193
|
Isolation and characterization of two fructokinase cDNA clones from rice
|
|
68194
|
Dynamically-driven enhancement of the catalytic machinery of the SARS 3C-like protease by the S284-T285-I286/A ...
|
|
68195
|
Dynamically-driven enhancement of the catalytic machinery of the SARS 3C-like protease by the S284-T285-I286/A ...
|
|
68196
|
Peptide aldehyde inhibitors challenge the substrate specificity of the SARS- coronavirus main protease
|
|
68197
|
Peptide aldehyde inhibitors challenge the substrate specificity of the SARS- coronavirus main protease
|
|
68198
|
Peptide aldehyde inhibitors challenge the substrate specificity of the SARS- coronavirus main protease
|
|
68199
|
Peptide aldehyde inhibitors challenge the substrate specificity of the SARS- coronavirus main protease
|
|
68200
|
Peptide aldehyde inhibitors challenge the substrate specificity of the SARS- coronavirus main protease
|
|
68201
|
Peptide aldehyde inhibitors challenge the substrate specificity of the SARS- coronavirus main protease
|
|
68202
|
Peptide aldehyde inhibitors challenge the substrate specificity of the SARS- coronavirus main protease
|
|
68203
|
Evidence that feedback inhibition of NAD kinase controls responses to oxidative stress
|
|
68204
|
Evidence that feedback inhibition of NAD kinase controls responses to oxidative stress
|
|
68205
|
Evidence that feedback inhibition of NAD kinase controls responses to oxidative stress
|
|
68206
|
Evidence that feedback inhibition of NAD kinase controls responses to oxidative stress
|
|
68207
|
Evidence that feedback inhibition of NAD kinase controls responses to oxidative stress
|
|
68208
|
Evidence that feedback inhibition of NAD kinase controls responses to oxidative stress
|
|
68209
|
Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, ...
|
|
68210
|
Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, ...
|
|
68211
|
Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, ...
|
|
68212
|
Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, ...
|
|
68213
|
Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, ...
|
|
68214
|
Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, ...
|
|
68215
|
Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, ...
|
|
68216
|
Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, ...
|
|
68217
|
Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, ...
|
|
68218
|
Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, ...
|
|
68219
|
Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, ...
|
|
68220
|
Structure-based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors ...
|
|
68221
|
Structure-based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors ...
|
|
68222
|
Structure-based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors ...
|
|
68223
|
Structure-based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors ...
|
|
68224
|
Structure-based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors ...
|
|
68225
|
Structure-based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors ...
|
|
68226
|
Structure-based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors ...
|
|
68227
|
Structure-based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors ...
|
|
68228
|
Structure-based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors ...
|
|
68229
|
Structure-based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors ...
|
|
68230
|
Structure-based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors ...
|
|
68231
|
Structure-based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors ...
|
|
68232
|
Structure-based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors ...
|
|
68233
|
Structure-based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors ...
|
|
68234
|
Structure-based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors ...
|
|
68235
|
Structure-based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors ...
|
|
68236
|
Structure-based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors ...
|
|
68237
|
Structure-based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors ...
|
|
68238
|
Structure-based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors ...
|
|
68239
|
Structure-based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors ...
|
|
68240
|
A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication
|
|
68241
|
A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication
|
|
68242
|
A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication
|
|
68243
|
A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication
|
|
68244
|
A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication
|
|
68245
|
A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication
|
|
68246
|
A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication
|
|
68247
|
A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication
|
|
68248
|
Glycerol-insensitive Arabidopsis mutants: gli1 seedlings lack glycerol kinase, accumulate glycerol and are ...
|
|
68249
|
Characterization of the 2'-5'-oligoadenylate synthetase ubiquitin-like family.
|
|
68250
|
Characterization of the 2'-5'-oligoadenylate synthetase ubiquitin-like family.
|
|
68251
|
Characterization of the 2'-5'-oligoadenylate synthetase ubiquitin-like family.
|
|
68252
|
Characterization of the 2'-5'-oligoadenylate synthetase ubiquitin-like family.
|
|
68253
|
Characterization of the 2'-5'-oligoadenylate synthetase ubiquitin-like family.
|
|
68254
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68255
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68256
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68257
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68258
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68259
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68260
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68261
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68262
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68263
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68264
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68265
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68266
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68267
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68268
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68269
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68270
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68271
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68272
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68273
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68274
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68275
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68276
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68277
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68278
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68279
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68280
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68281
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68282
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68283
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68284
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68285
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68286
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68287
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68288
|
X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human ...
|
|
68289
|
Identification and characterization of human ethanolaminephosphotransferase1
|
|
68290
|
Phosphoenolpyruvate synthetase from the hyperthermophilic archaeon Pyrococcus furiosus
|
|
68291
|
Phosphoenolpyruvate synthetase from the hyperthermophilic archaeon Pyrococcus furiosus
|
|
68292
|
Phosphoenolpyruvate synthetase from the hyperthermophilic archaeon Pyrococcus furiosus
|
|
68293
|
Phosphoenolpyruvate synthetase from the hyperthermophilic archaeon Pyrococcus furiosus
|
|
68294
|
Phosphoenolpyruvate synthetase from the hyperthermophilic archaeon Pyrococcus furiosus
|
|
68295
|
Identification of novel drug scaffolds for inhibition of SARS-CoV 3-Chymotrypsin-like protease using virtual ...
|
|
68296
|
Identification of novel drug scaffolds for inhibition of SARS-CoV 3-Chymotrypsin-like protease using virtual ...
|
|
68297
|
Identification of novel drug scaffolds for inhibition of SARS-CoV 3-Chymotrypsin-like protease using virtual ...
|
|
68298
|
Identification of novel drug scaffolds for inhibition of SARS-CoV 3-Chymotrypsin-like protease using virtual ...
|
|
68299
|
Identification of novel drug scaffolds for inhibition of SARS-CoV 3-Chymotrypsin-like protease using virtual ...
|
|
68300
|
Identification of novel drug scaffolds for inhibition of SARS-CoV 3-Chymotrypsin-like protease using virtual ...
|
|
68301
|
Identification of novel drug scaffolds for inhibition of SARS-CoV 3-Chymotrypsin-like protease using virtual ...
|
|
68302
|
Identification of novel drug scaffolds for inhibition of SARS-CoV 3-Chymotrypsin-like protease using virtual ...
|
|
68303
|
Identification of novel drug scaffolds for inhibition of SARS-CoV 3-Chymotrypsin-like protease using virtual ...
|
|
68304
|
Identification of novel drug scaffolds for inhibition of SARS-CoV 3-Chymotrypsin-like protease using virtual ...
|
|
68305
|
Identification of novel drug scaffolds for inhibition of SARS-CoV 3-Chymotrypsin-like protease using virtual ...
|
|
68306
|
Identification of novel drug scaffolds for inhibition of SARS-CoV 3-Chymotrypsin-like protease using virtual ...
|
|
68307
|
Identification of novel drug scaffolds for inhibition of SARS-CoV 3-Chymotrypsin-like protease using virtual ...
|
|
68308
|
Identification of novel drug scaffolds for inhibition of SARS-CoV 3-Chymotrypsin-like protease using virtual ...
|
|
68309
|
Identification of novel drug scaffolds for inhibition of SARS-CoV 3-Chymotrypsin-like protease using virtual ...
|
|
68310
|
Identification of novel drug scaffolds for inhibition of SARS-CoV 3-Chymotrypsin-like protease using virtual ...
|
|
68311
|
Identification of novel drug scaffolds for inhibition of SARS-CoV 3-Chymotrypsin-like protease using virtual ...
|
|
68312
|
Identification of novel drug scaffolds for inhibition of SARS-CoV 3-Chymotrypsin-like protease using virtual ...
|
|
68313
|
Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV. Version 3
|
|
68314
|
Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV. Version 3
|
|
68315
|
Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV. Version 3
|
|
68316
|
Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV. Version 3
|
|
68317
|
Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV. Version 3
|
|
68318
|
Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV. Version 3
|
|
68319
|
Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV. Version 3
|
|
68320
|
Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV. Version 3
|
|
68321
|
Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV. Version 3
|
|
68322
|
Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV. Version 3
|
|
68323
|
Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV. Version 3
|
|
68324
|
Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV. Version 3
|
|
68325
|
Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV. Version 3
|
|
68326
|
Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV. Version 3
|
|
68327
|
Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV. Version 3
|
|
68328
|
Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV. Version 3
|
|
68329
|
Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV. Version 3
|
|
68330
|
Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV. Version 3
|
|
68331
|
Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV. Version 3
|
|
68332
|
Identification and design of novel small molecule inhibitors against MERS-CoV papain-like protease via ...
|
|
68333
|
Identification and design of novel small molecule inhibitors against MERS-CoV papain-like protease via ...
|
|
68334
|
Identification and design of novel small molecule inhibitors against MERS-CoV papain-like protease via ...
|
|
68335
|
Identification and design of novel small molecule inhibitors against MERS-CoV papain-like protease via ...
|
|
68336
|
Identification and design of novel small molecule inhibitors against MERS-CoV papain-like protease via ...
|
|
68337
|
Identification and design of novel small molecule inhibitors against MERS-CoV papain-like protease via ...
|
|
68338
|
Identification and design of novel small molecule inhibitors against MERS-CoV papain-like protease via ...
|
|
68339
|
Identification and design of novel small molecule inhibitors against MERS-CoV papain-like protease via ...
|
|
68340
|
Identification and design of novel small molecule inhibitors against MERS-CoV papain-like protease via ...
|
|
68341
|
Identification and design of novel small molecule inhibitors against MERS-CoV papain-like protease via ...
|
|
68342
|
Identification and design of novel small molecule inhibitors against MERS-CoV papain-like protease via ...
|
|
68343
|
Identification and design of novel small molecule inhibitors against MERS-CoV papain-like protease via ...
|
|
68344
|
Identification and design of novel small molecule inhibitors against MERS-CoV papain-like protease via ...
|
|
68345
|
Identification and design of novel small molecule inhibitors against MERS-CoV papain-like protease via ...
|
|
68346
|
Identification and design of novel small molecule inhibitors against MERS-CoV papain-like protease via ...
|
|
68347
|
Identification and design of novel small molecule inhibitors against MERS-CoV papain-like protease via ...
|
|
68348
|
Identification and design of novel small molecule inhibitors against MERS-CoV papain-like protease via ...
|
|
68349
|
Identification and design of novel small molecule inhibitors against MERS-CoV papain-like protease via ...
|
|
68350
|
Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia ...
|
|
68351
|
Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia ...
|
|
68352
|
Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia ...
|
|
68353
|
Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia ...
|
|
68354
|
Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia ...
|
|
68355
|
Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia ...
|
|
68356
|
Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia ...
|
|
68357
|
Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia ...
|
|
68358
|
Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia ...
|
|
68359
|
Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia ...
|
|
68360
|
Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia ...
|
|
68361
|
Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia ...
|
|
68362
|
Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia ...
|
|
68363
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68364
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68365
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68366
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68367
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68368
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68369
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68370
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68371
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68372
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68373
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68374
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68375
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68376
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68377
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68378
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68379
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68380
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68381
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68382
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68383
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68384
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68385
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68386
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68387
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68388
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68389
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68390
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68391
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68392
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68393
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68394
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68395
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68396
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68397
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68398
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68399
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68400
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68401
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68402
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68403
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68404
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68405
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68406
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68407
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68408
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68409
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68410
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68411
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68412
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68413
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68414
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68415
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68416
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68417
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68418
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68419
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68420
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68421
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68422
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68423
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68424
|
α-Ketoamides as Broad- Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, ...
|
|
68425
|
Synthetic capacity of Arabidopsis phosphatidylinositol synthase 1 expressed in Escherichia coli
|
|
68426
|
Synthetic capacity of Arabidopsis phosphatidylinositol synthase 1 expressed in Escherichia coli
|
|
68427
|
Synthetic capacity of Arabidopsis phosphatidylinositol synthase 1 expressed in Escherichia coli
|
|
68428
|
Repression of Escherichia coli PhoP-PhoQ signaling by acetate reveals a regulatory role for acetyl coenzyme A
|
|
68429
|
Repression of Escherichia coli PhoP-PhoQ signaling by acetate reveals a regulatory role for acetyl coenzyme A
|
|
68430
|
Repression of Escherichia coli PhoP-PhoQ signaling by acetate reveals a regulatory role for acetyl coenzyme A
|
|
68431
|
Repression of Escherichia coli PhoP-PhoQ signaling by acetate reveals a regulatory role for acetyl coenzyme A
|
|
68432
|
Saturation mutagenesis reveals that GLU54 of norovirus 3C-like protease is not essential for the proteolytic ...
|
|
68433
|
Saturation mutagenesis reveals that GLU54 of norovirus 3C-like protease is not essential for the proteolytic ...
|
|
68434
|
Saturation mutagenesis reveals that GLU54 of norovirus 3C-like protease is not essential for the proteolytic ...
|
|
68435
|
Saturation mutagenesis reveals that GLU54 of norovirus 3C-like protease is not essential for the proteolytic ...
|
|
68436
|
Saturation mutagenesis reveals that GLU54 of norovirus 3C-like protease is not essential for the proteolytic ...
|
|
68437
|
Saturation mutagenesis reveals that GLU54 of norovirus 3C-like protease is not essential for the proteolytic ...
|
|
68438
|
Saturation mutagenesis reveals that GLU54 of norovirus 3C-like protease is not essential for the proteolytic ...
|
|
68439
|
Saturation mutagenesis reveals that GLU54 of norovirus 3C-like protease is not essential for the proteolytic ...
|
|
68440
|
Saturation mutagenesis reveals that GLU54 of norovirus 3C-like protease is not essential for the proteolytic ...
|
|
68441
|
Saturation mutagenesis reveals that GLU54 of norovirus 3C-like protease is not essential for the proteolytic ...
|
|
68442
|
Saturation mutagenesis reveals that GLU54 of norovirus 3C-like protease is not essential for the proteolytic ...
|
|
68443
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68444
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68445
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68446
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68447
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68448
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68449
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68450
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68451
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68452
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68453
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68454
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68455
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68456
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68457
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68458
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68459
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68460
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68461
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68462
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68463
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68464
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68465
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68466
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68467
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68468
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68469
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68470
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68471
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68472
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68473
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68474
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68475
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68476
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68477
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68478
|
Two NAD+-isocitrate dehydrogenase forms in Phycomyces blakesleeanus. Induction in response to acetate growth ...
|
|
68479
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68480
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68481
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68482
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68483
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68484
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68485
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68486
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68487
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68488
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68489
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68490
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68491
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68492
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68493
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68494
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68495
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68496
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68497
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68498
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68499
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68500
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68501
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68502
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68503
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68504
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68505
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68506
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68507
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68508
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68509
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68510
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68511
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68512
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68513
|
Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases
|
|
68514
|
Regulation of glutamate metabolism by protein kinases in mycobacteria.
|
|
68515
|
Regulation of glutamate metabolism by protein kinases in mycobacteria.
|
|
68516
|
Regulation of glutamate metabolism by protein kinases in mycobacteria.
|
|
68517
|
Regulation of glutamate metabolism by protein kinases in mycobacteria.
|
|
68518
|
Regulation of glutamate metabolism by protein kinases in mycobacteria.
|
|
68519
|
Two different dihydroorotate dehydrogenases from yeast Saccharomyces kluyveri
|
|
68520
|
Two different dihydroorotate dehydrogenases from yeast Saccharomyces kluyveri
|
|
68521
|
Two different dihydroorotate dehydrogenases from yeast Saccharomyces kluyveri
|
|
68522
|
Two different dihydroorotate dehydrogenases from yeast Saccharomyces kluyveri
|
|
68523
|
Two different dihydroorotate dehydrogenases from yeast Saccharomyces kluyveri
|
|
68524
|
Two different dihydroorotate dehydrogenases from yeast Saccharomyces kluyveri
|
|
68525
|
Two different dihydroorotate dehydrogenases from yeast Saccharomyces kluyveri
|
|
68526
|
Two different dihydroorotate dehydrogenases from yeast Saccharomyces kluyveri
|
|
68527
|
Two different dihydroorotate dehydrogenases from yeast Saccharomyces kluyveri
|
|
68528
|
Two different dihydroorotate dehydrogenases from yeast Saccharomyces kluyveri
|
|
68529
|
Two different dihydroorotate dehydrogenases from yeast Saccharomyces kluyveri
|
|
68530
|
Two different dihydroorotate dehydrogenases from yeast Saccharomyces kluyveri
|
|
68531
|
Two different dihydroorotate dehydrogenases from yeast Saccharomyces kluyveri
|
|
68532
|
Two different dihydroorotate dehydrogenases from yeast Saccharomyces kluyveri
|
|
68533
|
Purification and biochemical characterization of a hydroxyneurosporene desaturase involved in the biosynthetic ...
|
|
68534
|
The obligate intracellular parasite Toxoplasma gondii secretes a soluble phosphatidylserine decarboxylase
|
|
68535
|
Structural study reveals that Ser-354 determines substrate specificity on human histidine decarboxylase
|
|
68536
|
Structural study reveals that Ser-354 determines substrate specificity on human histidine decarboxylase
|
|
68537
|
Structural study reveals that Ser-354 determines substrate specificity on human histidine decarboxylase
|
|
68538
|
Structural study reveals that Ser-354 determines substrate specificity on human histidine decarboxylase
|
|
68539
|
Structural study reveals that Ser-354 determines substrate specificity on human histidine decarboxylase
|
|
68540
|
An alternative polyamine biosynthetic pathway is widespread in bacteria and essential for biofilm formation in ...
|
|
68541
|
An alternative polyamine biosynthetic pathway is widespread in bacteria and essential for biofilm formation in ...
|
|
68542
|
An alternative polyamine biosynthetic pathway is widespread in bacteria and essential for biofilm formation in ...
|
|
68543
|
An alternative polyamine biosynthetic pathway is widespread in bacteria and essential for biofilm formation in ...
|
|
68544
|
Ethylmalonyl-CoA decarboxylase, a new enzyme involved in metabolite proofreading
|
|
68545
|
Ethylmalonyl-CoA decarboxylase, a new enzyme involved in metabolite proofreading
|
|
68546
|
Ethylmalonyl-CoA decarboxylase, a new enzyme involved in metabolite proofreading
|
|
68547
|
Ethylmalonyl-CoA decarboxylase, a new enzyme involved in metabolite proofreading
|
|
68548
|
Structure of Main Protease from Human Coronavirus NL63: Insights for Wide Spectrum Anti-Coronavirus Drug ...
|
|
68549
|
Structure of Main Protease from Human Coronavirus NL63: Insights for Wide Spectrum Anti-Coronavirus Drug ...
|
|
68550
|
Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage ...
|
|
68551
|
Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage ...
|
|
68552
|
Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage ...
|
|
68553
|
Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage ...
|
|
68554
|
Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage ...
|
|
68555
|
Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage ...
|
|
68556
|
Role of glutamate decarboxylase-like protein 1 (GADL1) in taurine biosynthesis
|
|
68557
|
Role of glutamate decarboxylase-like protein 1 (GADL1) in taurine biosynthesis
|
|
68558
|
Identification and characterization of a re-citrate synthase in Dehalococcoides strain CBDB1
|
|
68559
|
Identification and characterization of a re-citrate synthase in Dehalococcoides strain CBDB1
|
|
68560
|
Purification in an active state and properties of the 3-step phytoene desaturase from Rhodobacter capsulatus ...
|
|
68561
|
Purification in an active state and properties of the 3-step phytoene desaturase from Rhodobacter capsulatus ...
|
|
68562
|
Purification in an active state and properties of the 3-step phytoene desaturase from Rhodobacter capsulatus ...
|
|
68563
|
Purification in an active state and properties of the 3-step phytoene desaturase from Rhodobacter capsulatus ...
|
|
68564
|
Phytoene desaturase: heterologous expression in an active state, purification, and biochemical properties
|
|
68565
|
Phytoene desaturase: heterologous expression in an active state, purification, and biochemical properties
|
|
68566
|
Inhibition of a Putative Dihydropyrimidinase from Pseudomonas aeruginosa PAO1 by Flavonoids and Substrates of ...
|
|
68567
|
Inhibition of a Putative Dihydropyrimidinase from Pseudomonas aeruginosa PAO1 by Flavonoids and Substrates of ...
|
|
68568
|
Inhibition of a Putative Dihydropyrimidinase from Pseudomonas aeruginosa PAO1 by Flavonoids and Substrates of ...
|
|
68569
|
Inhibition of a Putative Dihydropyrimidinase from Pseudomonas aeruginosa PAO1 by Flavonoids and Substrates of ...
|
|
68570
|
Inhibition of a Putative Dihydropyrimidinase from Pseudomonas aeruginosa PAO1 by Flavonoids and Substrates of ...
|
|
68571
|
Inhibition of a Putative Dihydropyrimidinase from Pseudomonas aeruginosa PAO1 by Flavonoids and Substrates of ...
|
|
68572
|
Inhibition of a Putative Dihydropyrimidinase from Pseudomonas aeruginosa PAO1 by Flavonoids and Substrates of ...
|
|
68573
|
Inhibition of a Putative Dihydropyrimidinase from Pseudomonas aeruginosa PAO1 by Flavonoids and Substrates of ...
|
|
68574
|
Inhibition of a Putative Dihydropyrimidinase from Pseudomonas aeruginosa PAO1 by Flavonoids and Substrates of ...
|
|
68575
|
Inhibition of a Putative Dihydropyrimidinase from Pseudomonas aeruginosa PAO1 by Flavonoids and Substrates of ...
|
|
68576
|
Inhibition of a Putative Dihydropyrimidinase from Pseudomonas aeruginosa PAO1 by Flavonoids and Substrates of ...
|
|
68577
|
Inhibition of a Putative Dihydropyrimidinase from Pseudomonas aeruginosa PAO1 by Flavonoids and Substrates of ...
|
|
68578
|
Inhibition of a Putative Dihydropyrimidinase from Pseudomonas aeruginosa PAO1 by Flavonoids and Substrates of ...
|
|
68579
|
Inhibition of a Putative Dihydropyrimidinase from Pseudomonas aeruginosa PAO1 by Flavonoids and Substrates of ...
|
|
68580
|
Inhibition of a Putative Dihydropyrimidinase from Pseudomonas aeruginosa PAO1 by Flavonoids and Substrates of ...
|
|
68581
|
Inhibition of a Putative Dihydropyrimidinase from Pseudomonas aeruginosa PAO1 by Flavonoids and Substrates of ...
|
|
68582
|
Inhibition of a Putative Dihydropyrimidinase from Pseudomonas aeruginosa PAO1 by Flavonoids and Substrates of ...
|
|
68583
|
Inhibition of a Putative Dihydropyrimidinase from Pseudomonas aeruginosa PAO1 by Flavonoids and Substrates of ...
|
|
68584
|
Inhibition of a Putative Dihydropyrimidinase from Pseudomonas aeruginosa PAO1 by Flavonoids and Substrates of ...
|
|
68585
|
Inhibition of a Putative Dihydropyrimidinase from Pseudomonas aeruginosa PAO1 by Flavonoids and Substrates of ...
|
|
68586
|
Mouse steroid sulfotransferases: substrate specificity and preliminary X-ray crystallographic analysis
|
|
68587
|
Mouse steroid sulfotransferases: substrate specificity and preliminary X-ray crystallographic analysis
|
|
68588
|
Mouse steroid sulfotransferases: substrate specificity and preliminary X-ray crystallographic analysis
|
|
68589
|
Mouse steroid sulfotransferases: substrate specificity and preliminary X-ray crystallographic analysis
|
|
68590
|
Mouse steroid sulfotransferases: substrate specificity and preliminary X-ray crystallographic analysis
|
|
68591
|
Mouse steroid sulfotransferases: substrate specificity and preliminary X-ray crystallographic analysis
|
|
68592
|
Mouse steroid sulfotransferases: substrate specificity and preliminary X-ray crystallographic analysis
|
|
68593
|
Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes
|
|
68594
|
Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes
|
|
68595
|
Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes
|
|
68596
|
Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes
|
|
68597
|
Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes
|
|
68598
|
Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes
|
|
68599
|
Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes
|
|
68600
|
Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes
|
|
68601
|
Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes
|
|
68602
|
Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes
|
|
68603
|
Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes
|
|
68604
|
Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes
|
|
68605
|
Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes
|
|
68606
|
Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes
|
|
68607
|
Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes
|
|
68608
|
Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes
|
|
68609
|
Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes
|
|
68610
|
Disulfiram can inhibit MERS and SARS coronavirus papain-like proteases via different modes
|
|
68611
|
Porcine epidemic diarrhea virus papain-like protease 2 can be noncompetitively inhibited by 6-thioguanine
|
|
68612
|
Porcine epidemic diarrhea virus papain-like protease 2 can be noncompetitively inhibited by 6-thioguanine
|
|
68613
|
Porcine epidemic diarrhea virus papain-like protease 2 can be noncompetitively inhibited by 6-thioguanine
|
|
68614
|
Porcine epidemic diarrhea virus papain-like protease 2 can be noncompetitively inhibited by 6-thioguanine
|
|
68615
|
Porcine epidemic diarrhea virus papain-like protease 2 can be noncompetitively inhibited by 6-thioguanine
|
|
68616
|
Porcine epidemic diarrhea virus papain-like protease 2 can be noncompetitively inhibited by 6-thioguanine
|
|
68617
|
Porcine epidemic diarrhea virus papain-like protease 2 can be noncompetitively inhibited by 6-thioguanine
|
|
68618
|
Thiopurine analogues inhibit papain-like protease of severe acute respiratory syndrome coronavirus
|
|
68619
|
Thiopurine analogues inhibit papain-like protease of severe acute respiratory syndrome coronavirus
|
|
68620
|
Thiopurine analogues inhibit papain-like protease of severe acute respiratory syndrome coronavirus
|
|
68621
|
Thiopurine analogues inhibit papain-like protease of severe acute respiratory syndrome coronavirus
|
|
68622
|
Thiopurine analogues inhibit papain-like protease of severe acute respiratory syndrome coronavirus
|
|
68623
|
Thiopurine analogues inhibit papain-like protease of severe acute respiratory syndrome coronavirus
|
|
68624
|
Thiopurine analogues inhibit papain-like protease of severe acute respiratory syndrome coronavirus
|
|
68625
|
Thiopurine analogues inhibit papain-like protease of severe acute respiratory syndrome coronavirus
|
|
68626
|
Thiopurine analogues inhibit papain-like protease of severe acute respiratory syndrome coronavirus
|
|
68627
|
Thiopurine analogues inhibit papain-like protease of severe acute respiratory syndrome coronavirus
|
|
68628
|
High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase
|
|
68629
|
High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase
|
|
68630
|
High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase
|
|
68631
|
High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase
|
|
68632
|
High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase
|
|
68633
|
High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase
|
|
68634
|
High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase
|
|
68635
|
High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase
|
|
68636
|
High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase
|
|
68637
|
High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase
|
|
68638
|
Thiopurine analogs and mycophenolic acid synergistically inhibit the papain-like protease of Middle East ...
|
|
68639
|
Thiopurine analogs and mycophenolic acid synergistically inhibit the papain-like protease of Middle East ...
|
|
68640
|
Thiopurine analogs and mycophenolic acid synergistically inhibit the papain-like protease of Middle East ...
|
|
68641
|
Thiopurine analogs and mycophenolic acid synergistically inhibit the papain-like protease of Middle East ...
|
|
68642
|
Thiopurine analogs and mycophenolic acid synergistically inhibit the papain-like protease of Middle East ...
|
|
68643
|
Thiopurine analogs and mycophenolic acid synergistically inhibit the papain-like protease of Middle East ...
|
|
68644
|
Thiopurine analogs and mycophenolic acid synergistically inhibit the papain-like protease of Middle East ...
|
|
68645
|
Thiopurine analogs and mycophenolic acid synergistically inhibit the papain-like protease of Middle East ...
|
|
68646
|
Thiopurine analogs and mycophenolic acid synergistically inhibit the papain-like protease of Middle East ...
|
|
68647
|
Thiopurine analogs and mycophenolic acid synergistically inhibit the papain-like protease of Middle East ...
|
|
68648
|
Thiopurine analogs and mycophenolic acid synergistically inhibit the papain-like protease of Middle East ...
|
|
68649
|
X-ray Structure and Enzymatic Activity Profile of a Core Papain-like Protease of MERS Coronavirus with utility ...
|
|
68650
|
X-ray Structure and Enzymatic Activity Profile of a Core Papain-like Protease of MERS Coronavirus with utility ...
|
|
68651
|
X-ray Structure and Enzymatic Activity Profile of a Core Papain-like Protease of MERS Coronavirus with utility ...
|
|
68652
|
X-ray Structure and Enzymatic Activity Profile of a Core Papain-like Protease of MERS Coronavirus with utility ...
|
|
68653
|
X-ray Structure and Enzymatic Activity Profile of a Core Papain-like Protease of MERS Coronavirus with utility ...
|
|
68654
|
X-ray Structure and Enzymatic Activity Profile of a Core Papain-like Protease of MERS Coronavirus with utility ...
|
|
68655
|
X-ray Structure and Enzymatic Activity Profile of a Core Papain-like Protease of MERS Coronavirus with utility ...
|
|
68656
|
X-ray Structure and Enzymatic Activity Profile of a Core Papain-like Protease of MERS Coronavirus with utility ...
|
|
68657
|
X-ray Structure and Enzymatic Activity Profile of a Core Papain-like Protease of MERS Coronavirus with utility ...
|
|
68658
|
X-ray Structure and Enzymatic Activity Profile of a Core Papain-like Protease of MERS Coronavirus with utility ...
|
|
68659
|
X-ray Structure and Enzymatic Activity Profile of a Core Papain-like Protease of MERS Coronavirus with utility ...
|
|
68660
|
X-ray Structure and Enzymatic Activity Profile of a Core Papain-like Protease of MERS Coronavirus with utility ...
|
|
68661
|
X-ray Structure and Enzymatic Activity Profile of a Core Papain-like Protease of MERS Coronavirus with utility ...
|
|
68662
|
X-ray Structure and Enzymatic Activity Profile of a Core Papain-like Protease of MERS Coronavirus with utility ...
|
|
68663
|
X-ray Structure and Enzymatic Activity Profile of a Core Papain-like Protease of MERS Coronavirus with utility ...
|
|
68664
|
X-ray Structure and Enzymatic Activity Profile of a Core Papain-like Protease of MERS Coronavirus with utility ...
|
|
68665
|
Conservation of the hydroxysteroid sulfotransferase SULT2B1 gene structure in the mouse: pre- and postnatal ...
|
|
68666
|
Conservation of the hydroxysteroid sulfotransferase SULT2B1 gene structure in the mouse: pre- and postnatal ...
|
|
68667
|
Conservation of the hydroxysteroid sulfotransferase SULT2B1 gene structure in the mouse: pre- and postnatal ...
|
|
68668
|
Conservation of the hydroxysteroid sulfotransferase SULT2B1 gene structure in the mouse: pre- and postnatal ...
|
|
68669
|
Conservation of the hydroxysteroid sulfotransferase SULT2B1 gene structure in the mouse: pre- and postnatal ...
|
|
68670
|
Conservation of the hydroxysteroid sulfotransferase SULT2B1 gene structure in the mouse: pre- and postnatal ...
|
|
68671
|
Conservation of the hydroxysteroid sulfotransferase SULT2B1 gene structure in the mouse: pre- and postnatal ...
|
|
68672
|
Conservation of the hydroxysteroid sulfotransferase SULT2B1 gene structure in the mouse: pre- and postnatal ...
|
|
68673
|
Geranyl diphosphate:4-hydroxybenzoate geranyltransferase from Lithospermum erythrorhizon. Cloning and ...
|
|
68674
|
Geranyl diphosphate:4-hydroxybenzoate geranyltransferase from Lithospermum erythrorhizon. Cloning and ...
|
|
68675
|
Geranyl diphosphate:4-hydroxybenzoate geranyltransferase from Lithospermum erythrorhizon. Cloning and ...
|
|
68676
|
Geranyl diphosphate:4-hydroxybenzoate geranyltransferase from Lithospermum erythrorhizon. Cloning and ...
|
|
68677
|
Crystal structures of ADP and AMPPNP-bound propionate kinase (TdcD) from Salmonella typhimurium: comparison ...
|
|
68678
|
Crystal structures of ADP and AMPPNP-bound propionate kinase (TdcD) from Salmonella typhimurium: comparison ...
|
|
68679
|
Crystal structures of ADP and AMPPNP-bound propionate kinase (TdcD) from Salmonella typhimurium: comparison ...
|
|
68680
|
Regulation of the nonreceptor tyrosine kinase Brk by autophosphorylation and by autoinhibition
|
|
68681
|
Regulation of the nonreceptor tyrosine kinase Brk by autophosphorylation and by autoinhibition
|
|
68682
|
Regulation of the nonreceptor tyrosine kinase Brk by autophosphorylation and by autoinhibition
|
|
68683
|
Regulation of the nonreceptor tyrosine kinase Brk by autophosphorylation and by autoinhibition
|
|
68684
|
Regulation of the nonreceptor tyrosine kinase Brk by autophosphorylation and by autoinhibition
|
|
68685
|
Long-range cooperative interactions modulate dimerization in SARS 3CLpro
|
|
68686
|
Long-range cooperative interactions modulate dimerization in SARS 3CLpro
|
|
68687
|
Long-range cooperative interactions modulate dimerization in SARS 3CLpro
|
|
68688
|
Long-range cooperative interactions modulate dimerization in SARS 3CLpro
|
|
68689
|
Long-range cooperative interactions modulate dimerization in SARS 3CLpro
|
|
68690
|
Long-range cooperative interactions modulate dimerization in SARS 3CLpro
|
|
68691
|
Long-range cooperative interactions modulate dimerization in SARS 3CLpro
|
|
68692
|
Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding ...
|
|
68693
|
Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding ...
|
|
68694
|
Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding ...
|
|
68695
|
Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding ...
|
|
68696
|
Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding ...
|
|
68697
|
Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding ...
|
|
68698
|
Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding ...
|
|
68699
|
Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding ...
|
|
68700
|
Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding ...
|
|
68701
|
Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding ...
|
|
68702
|
Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding ...
|
|
68703
|
Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding ...
|
|
68704
|
Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding ...
|
|
68705
|
Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding ...
|
|
68706
|
Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding ...
|
|
68707
|
Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding ...
|
|
68708
|
Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding ...
|
|
68709
|
Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding ...
|
|
68710
|
Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding ...
|
|
68711
|
Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding ...
|
|
68712
|
Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding ...
|
|
68713
|
Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding ...
|
|
68714
|
Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding ...
|
|
68715
|
Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding ...
|
|
68716
|
Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding ...
|
|
68717
|
Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding ...
|
|
68718
|
Biosynthesis of pteridines in Escherichia coli. Structural and mechanistic similarity of ...
|
|
68719
|
Biosynthesis of pteridines in Escherichia coli. Structural and mechanistic similarity of ...
|
|
68720
|
Biosynthesis of pteridines in Escherichia coli. Structural and mechanistic similarity of ...
|
|
68721
|
Biosynthesis of pteridines in Escherichia coli. Structural and mechanistic similarity of ...
|
|
68722
|
Biosynthesis of pteridines in Escherichia coli. Structural and mechanistic similarity of ...
|
|
68723
|
Biosynthesis of pteridines in Escherichia coli. Structural and mechanistic similarity of ...
|
|
68724
|
Biosynthesis of pteridines in Escherichia coli. Structural and mechanistic similarity of ...
|
|
68725
|
Biosynthesis of pteridines in Escherichia coli. Structural and mechanistic similarity of ...
|
|
68726
|
Biosynthesis of pteridines in Escherichia coli. Structural and mechanistic similarity of ...
|
|
68727
|
Biosynthesis of pteridines in Escherichia coli. Structural and mechanistic similarity of ...
|
|
68728
|
Biosynthesis of pteridines in Escherichia coli. Structural and mechanistic similarity of ...
|
|
68729
|
Purification and properties of isocitrate dehydrogenase (NADP) from Thermus aquaticus YT-1, Bacillus ...
|
|
68730
|
Purification and properties of isocitrate dehydrogenase (NADP) from Thermus aquaticus YT-1, Bacillus ...
|
|
68731
|
Purification and properties of isocitrate dehydrogenase (NADP) from Thermus aquaticus YT-1, Bacillus ...
|
|
68732
|
Purification and properties of isocitrate dehydrogenase (NADP) from Thermus aquaticus YT-1, Bacillus ...
|
|
68733
|
Purification and properties of isocitrate dehydrogenase (NADP) from Thermus aquaticus YT-1, Bacillus ...
|
|
68734
|
Purification and properties of isocitrate dehydrogenase (NADP) from Thermus aquaticus YT-1, Bacillus ...
|
|
68735
|
Purification and properties of isocitrate dehydrogenase (NADP) from Thermus aquaticus YT-1, Bacillus ...
|
|
68736
|
Purification and properties of isocitrate dehydrogenase (NADP) from Thermus aquaticus YT-1, Bacillus ...
|
|
68737
|
Progress Curve Analysis Within BioCatNet: Comparing Kinetic Models for Enzyme-Catalyzed Self-Ligation
|
|
68738
|
Progress Curve Analysis Within BioCatNet: Comparing Kinetic Models for Enzyme-Catalyzed Self-Ligation
|
|
68739
|
Progress Curve Analysis Within BioCatNet: Comparing Kinetic Models for Enzyme-Catalyzed Self-Ligation
|
|
68740
|
Progress Curve Analysis Within BioCatNet: Comparing Kinetic Models for Enzyme-Catalyzed Self-Ligation
|
|
68741
|
Progress Curve Analysis Within BioCatNet: Comparing Kinetic Models for Enzyme-Catalyzed Self-Ligation
|
|
68742
|
Progress Curve Analysis Within BioCatNet: Comparing Kinetic Models for Enzyme-Catalyzed Self-Ligation
|
|
68743
|
Progress Curve Analysis Within BioCatNet: Comparing Kinetic Models for Enzyme-Catalyzed Self-Ligation
|
|
68744
|
Progress Curve Analysis Within BioCatNet: Comparing Kinetic Models for Enzyme-Catalyzed Self-Ligation
|
|
68745
|
Discovery of novel non- competitive inhibitors of mammalian neutral M1 aminopeptidase (APN)
|
|
68746
|
Discovery of novel non- competitive inhibitors of mammalian neutral M1 aminopeptidase (APN)
|
|
68747
|
Discovery of novel non- competitive inhibitors of mammalian neutral M1 aminopeptidase (APN)
|
|
68748
|
Discovery of novel non- competitive inhibitors of mammalian neutral M1 aminopeptidase (APN)
|
|
68749
|
Discovery of novel non- competitive inhibitors of mammalian neutral M1 aminopeptidase (APN)
|
|
68750
|
Discovery of novel non- competitive inhibitors of mammalian neutral M1 aminopeptidase (APN)
|
|
68751
|
Discovery of novel non- competitive inhibitors of mammalian neutral M1 aminopeptidase (APN)
|
|
68752
|
Discovery of novel non- competitive inhibitors of mammalian neutral M1 aminopeptidase (APN)
|
|
68753
|
Discovery of novel non- competitive inhibitors of mammalian neutral M1 aminopeptidase (APN)
|
|
68754
|
Discovery of novel non- competitive inhibitors of mammalian neutral M1 aminopeptidase (APN)
|
|
68755
|
Discovery of novel non- competitive inhibitors of mammalian neutral M1 aminopeptidase (APN)
|
|
68756
|
Discovery of novel non- competitive inhibitors of mammalian neutral M1 aminopeptidase (APN)
|
|
68757
|
Discovery of novel non- competitive inhibitors of mammalian neutral M1 aminopeptidase (APN)
|
|
68758
|
Discovery of novel non- competitive inhibitors of mammalian neutral M1 aminopeptidase (APN)
|
|
68759
|
Discovery of novel non- competitive inhibitors of mammalian neutral M1 aminopeptidase (APN)
|
|
68760
|
Discovery of novel non- competitive inhibitors of mammalian neutral M1 aminopeptidase (APN)
|
|
68761
|
Discovery of novel non- competitive inhibitors of mammalian neutral M1 aminopeptidase (APN)
|
|
68762
|
Discovery of novel non- competitive inhibitors of mammalian neutral M1 aminopeptidase (APN)
|
|
68763
|
Discovery of novel non- competitive inhibitors of mammalian neutral M1 aminopeptidase (APN)
|
|
68764
|
Discovery of novel non- competitive inhibitors of mammalian neutral M1 aminopeptidase (APN)
|
|
68765
|
Cyclohexanecarboxyl-coenzyme A (CoA) and cyclohex-1-ene-1-carboxyl-CoA dehydrogenases, two enzymes involved in ...
|
|
68766
|
Cyclohexanecarboxyl-coenzyme A (CoA) and cyclohex-1-ene-1-carboxyl-CoA dehydrogenases, two enzymes involved in ...
|
|
68767
|
Cyclohexanecarboxyl-coenzyme A (CoA) and cyclohex-1-ene-1-carboxyl-CoA dehydrogenases, two enzymes involved in ...
|
|
68768
|
Cyclohexanecarboxyl-coenzyme A (CoA) and cyclohex-1-ene-1-carboxyl-CoA dehydrogenases, two enzymes involved in ...
|
|
68769
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68770
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68771
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68772
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68773
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68774
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68775
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68776
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68777
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68778
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68779
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68780
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68781
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68782
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68783
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68784
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68785
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68786
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68787
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68788
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68789
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68790
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68791
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68792
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68793
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68794
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68795
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68796
|
Discovery of potent and selective inhibitors of the Escherichia coli M1-aminopeptidase via multicomponent ...
|
|
68797
|
MGAT2, a monoacylglycerol acyltransferase expressed in the small intestine
|
|
68798
|
MGAT2, a monoacylglycerol acyltransferase expressed in the small intestine
|
|
68799
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68800
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68801
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68802
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68803
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68804
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68805
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68806
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68807
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68808
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68809
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68810
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68811
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68812
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68813
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68814
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68815
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68816
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68817
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68818
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68819
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68820
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68821
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68822
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68823
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68824
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68825
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68826
|
Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA(Cys)
|
|
68827
|
Characterization of the alpha-ketoglutarate dehydrogenase complex from Fasciola hepatica: potential ...
|
|
68828
|
Characterization of the alpha-ketoglutarate dehydrogenase complex from Fasciola hepatica: potential ...
|
|
68829
|
Characterization of the alpha-ketoglutarate dehydrogenase complex from Fasciola hepatica: potential ...
|
|
68830
|
Characterization of the alpha-ketoglutarate dehydrogenase complex from Fasciola hepatica: potential ...
|
|
68831
|
Characterization of the alpha-ketoglutarate dehydrogenase complex from Fasciola hepatica: potential ...
|
|
68832
|
Characterization of the alpha-ketoglutarate dehydrogenase complex from Fasciola hepatica: potential ...
|
|
68833
|
Characterization of the alpha-ketoglutarate dehydrogenase complex from Fasciola hepatica: potential ...
|
|
68834
|
Characterization of the alpha-ketoglutarate dehydrogenase complex from Fasciola hepatica: potential ...
|
|
68835
|
Characterization of the alpha-ketoglutarate dehydrogenase complex from Fasciola hepatica: potential ...
|
|
68836
|
Mutational analysis of a helicase motif-based RNA 5'-triphosphatase/NTPase from bamboo mosaic virus.
|
|
68837
|
Mutational analysis of a helicase motif-based RNA 5'-triphosphatase/NTPase from bamboo mosaic virus.
|
|
68838
|
Mutational analysis of a helicase motif-based RNA 5'-triphosphatase/NTPase from bamboo mosaic virus.
|
|
68839
|
Mutational analysis of a helicase motif-based RNA 5'-triphosphatase/NTPase from bamboo mosaic virus.
|
|
68840
|
Mutational analysis of a helicase motif-based RNA 5'-triphosphatase/NTPase from bamboo mosaic virus.
|
|
68841
|
Mutational analysis of a helicase motif-based RNA 5'-triphosphatase/NTPase from bamboo mosaic virus.
|
|
68842
|
Mutational analysis of a helicase motif-based RNA 5'-triphosphatase/NTPase from bamboo mosaic virus.
|
|
68843
|
Mutational analysis of a helicase motif-based RNA 5'-triphosphatase/NTPase from bamboo mosaic virus.
|
|
68844
|
Mutational analysis of a helicase motif-based RNA 5'-triphosphatase/NTPase from bamboo mosaic virus.
|
|
68845
|
Mutational analysis of a helicase motif-based RNA 5'-triphosphatase/NTPase from bamboo mosaic virus.
|
|
68846
|
Mutational analysis of a helicase motif-based RNA 5'-triphosphatase/NTPase from bamboo mosaic virus.
|
|
68847
|
Mutational analysis of a helicase motif-based RNA 5'-triphosphatase/NTPase from bamboo mosaic virus.
|
|
68848
|
Isolation and characterization of two tryptophan biosynthetic enzymes, indoleglycerol phosphate synthase and ...
|
|
68849
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68850
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68851
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68852
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68853
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68854
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68855
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68856
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68857
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68858
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68859
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68860
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68861
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68862
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68863
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68864
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68865
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68866
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68867
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68868
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68869
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68870
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68871
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68872
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68873
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68874
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68875
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68876
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68877
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68878
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68879
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68880
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68881
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68882
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68883
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68884
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68885
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68886
|
The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase ...
|
|
68887
|
Regulatory properties of a plant NAD: isocitrate dehydrogenase. The effect of inorganic ions.
|
|
68888
|
Regulatory properties of a plant NAD: isocitrate dehydrogenase. The effect of inorganic ions.
|
|
68889
|
Regulatory properties of a plant NAD: isocitrate dehydrogenase. The effect of inorganic ions.
|
|
68890
|
Regulatory properties of a plant NAD: isocitrate dehydrogenase. The effect of inorganic ions.
|
|
68891
|
Regulatory properties of a plant NAD: isocitrate dehydrogenase. The effect of inorganic ions.
|
|
68892
|
Regulatory properties of a plant NAD: isocitrate dehydrogenase. The effect of inorganic ions.
|
|
68893
|
Regulatory properties of a plant NAD: isocitrate dehydrogenase. The effect of inorganic ions.
|
|
68894
|
Regulatory properties of a plant NAD: isocitrate dehydrogenase. The effect of inorganic ions.
|
|
68895
|
Regulatory properties of a plant NAD: isocitrate dehydrogenase. The effect of inorganic ions.
|
|
68896
|
Regulatory properties of a plant NAD: isocitrate dehydrogenase. The effect of inorganic ions.
|
|
68897
|
Regulatory properties of a plant NAD: isocitrate dehydrogenase. The effect of inorganic ions.
|
|
68898
|
Regulatory properties of a plant NAD: isocitrate dehydrogenase. The effect of inorganic ions.
|
|
68899
|
Relationship of Catalysis and Active Site Loop Dynamics in the (βα)(8)-Barrel Enzyme Indole-3-glycerol ...
|
|
68900
|
Relationship of Catalysis and Active Site Loop Dynamics in the (βα)(8)-Barrel Enzyme Indole-3-glycerol ...
|
|
68901
|
Relationship of Catalysis and Active Site Loop Dynamics in the (βα)(8)-Barrel Enzyme Indole-3-glycerol ...
|
|
68902
|
Relationship of Catalysis and Active Site Loop Dynamics in the (βα)(8)-Barrel Enzyme Indole-3-glycerol ...
|
|
68903
|
Relationship of Catalysis and Active Site Loop Dynamics in the (βα)(8)-Barrel Enzyme Indole-3-glycerol ...
|
|
68904
|
Relationship of Catalysis and Active Site Loop Dynamics in the (βα)(8)-Barrel Enzyme Indole-3-glycerol ...
|
|
68905
|
Purification and characterization of the indole-3-glycerolphosphate synthase/anthranilate synthase complex of ...
|
|
68906
|
Purification and characterization of the indole-3-glycerolphosphate synthase/anthranilate synthase complex of ...
|
|
68907
|
Purification and characterization of the indole-3-glycerolphosphate synthase/anthranilate synthase complex of ...
|
|
68908
|
Purification and characterization of the indole-3-glycerolphosphate synthase/anthranilate synthase complex of ...
|
|
68909
|
Purification and characterization of the indole-3-glycerolphosphate synthase/anthranilate synthase complex of ...
|
|
68910
|
Purification and characterization of the indole-3-glycerolphosphate synthase/anthranilate synthase complex of ...
|
|
68911
|
Purification and characterization of the indole-3-glycerolphosphate synthase/anthranilate synthase complex of ...
|
|
68912
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68913
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68914
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68915
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68916
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68917
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68918
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68919
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68920
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68921
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68922
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68923
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68924
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68925
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68926
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68927
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68928
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68929
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68930
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68931
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68932
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68933
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68934
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68935
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68936
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68937
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68938
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68939
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68940
|
Purification and properties of diphosphopyridine nuleotide-linked isocitrate dehydrogenase of mammalian liver.
|
|
68941
|
A novel thermostable beta-galactosidase from Bacillus coagulans with excellent hydrolysis ability for lactose ...
|
|
68942
|
A novel thermostable beta-galactosidase from Bacillus coagulans with excellent hydrolysis ability for lactose ...
|
|
68943
|
A novel thermostable beta-galactosidase from Bacillus coagulans with excellent hydrolysis ability for lactose ...
|
|
68944
|
A novel thermostable beta-galactosidase from Bacillus coagulans with excellent hydrolysis ability for lactose ...
|
|
68945
|
Improvement of Selenomonas ruminantium β-xylosidase thermal stability by replacing buried free cysteines via ...
|
|
68946
|
Improvement of Selenomonas ruminantium β-xylosidase thermal stability by replacing buried free cysteines via ...
|
|
68947
|
Improvement of Selenomonas ruminantium β-xylosidase thermal stability by replacing buried free cysteines via ...
|
|
68948
|
Improvement of Selenomonas ruminantium β-xylosidase thermal stability by replacing buried free cysteines via ...
|
|
68949
|
Improvement of Selenomonas ruminantium β-xylosidase thermal stability by replacing buried free cysteines via ...
|
|
68950
|
Characterization of the indole-3-glycerol phosphate synthase from Pseudomonas aeruginosa PAO1
|
|
68951
|
2-Ketogluconate Kinase from Cupriavidus necator H16: Purification, Characterization, and Exploration of Its ...
|
|
68952
|
2-Ketogluconate Kinase from Cupriavidus necator H16: Purification, Characterization, and Exploration of Its ...
|
|
68953
|
2-Ketogluconate Kinase from Cupriavidus necator H16: Purification, Characterization, and Exploration of Its ...
|
|
68954
|
2-Ketogluconate Kinase from Cupriavidus necator H16: Purification, Characterization, and Exploration of Its ...
|
|
68955
|
2-Ketogluconate Kinase from Cupriavidus necator H16: Purification, Characterization, and Exploration of Its ...
|
|
68956
|
Differences in the catalytic mechanisms of mesophilic and thermophilic indole-3-glycerol phosphate synthase ...
|
|
68957
|
Differences in the catalytic mechanisms of mesophilic and thermophilic indole-3-glycerol phosphate synthase ...
|
|
68958
|
Differences in the catalytic mechanisms of mesophilic and thermophilic indole-3-glycerol phosphate synthase ...
|
|
68959
|
Differences in the catalytic mechanisms of mesophilic and thermophilic indole-3-glycerol phosphate synthase ...
|
|
68960
|
Differences in the catalytic mechanisms of mesophilic and thermophilic indole-3-glycerol phosphate synthase ...
|
|
68961
|
Differences in the catalytic mechanisms of mesophilic and thermophilic indole-3-glycerol phosphate synthase ...
|
|
68962
|
Comparison of the inhibition potential of parthenolide and micheliolide on various UDP-glucuronosyltransferase ...
|
|
68963
|
Single amino acid mutation altered substrate specificity for L-glucose and inositol in scyllo-inositol ...
|
|
68964
|
Single amino acid mutation altered substrate specificity for L-glucose and inositol in scyllo-inositol ...
|
|
68965
|
Single amino acid mutation altered substrate specificity for L-glucose and inositol in scyllo-inositol ...
|
|
68966
|
Single amino acid mutation altered substrate specificity for L-glucose and inositol in scyllo-inositol ...
|
|
68967
|
Single amino acid mutation altered substrate specificity for L-glucose and inositol in scyllo-inositol ...
|
|
68968
|
Single amino acid mutation altered substrate specificity for L-glucose and inositol in scyllo-inositol ...
|
|
68969
|
Single amino acid mutation altered substrate specificity for L-glucose and inositol in scyllo-inositol ...
|
|
68970
|
Single amino acid mutation altered substrate specificity for L-glucose and inositol in scyllo-inositol ...
|
|
68971
|
Overexpression, purification and biochemical characterization of the wound-induced leucine aminopeptidase of ...
|
|
68972
|
Overexpression, purification and biochemical characterization of the wound-induced leucine aminopeptidase of ...
|
|
68973
|
PIP4Kbeta interacts with and modulates nuclear localization of the high-activity PtdIns5P-4-kinase isoform ...
|
|
68974
|
PIP4Kbeta interacts with and modulates nuclear localization of the high-activity PtdIns5P-4-kinase isoform ...
|
|
68975
|
Metal binding stoichiometry and mechanism of metal ion modulation of the activity of porcine kidney leucine ...
|
|
68976
|
Metal binding stoichiometry and mechanism of metal ion modulation of the activity of porcine kidney leucine ...
|
|
68977
|
Metal binding stoichiometry and mechanism of metal ion modulation of the activity of porcine kidney leucine ...
|
|
68978
|
Metal binding stoichiometry and mechanism of metal ion modulation of the activity of porcine kidney leucine ...
|
|
68979
|
Metal binding stoichiometry and mechanism of metal ion modulation of the activity of porcine kidney leucine ...
|
|
68980
|
Metal binding stoichiometry and mechanism of metal ion modulation of the activity of porcine kidney leucine ...
|
|
68981
|
Metal binding stoichiometry and mechanism of metal ion modulation of the activity of porcine kidney leucine ...
|
|
68982
|
Purification and partial characterization of barley leucine aminopeptidase.
|
|
68983
|
Purification and partial characterization of barley leucine aminopeptidase.
|
|
68984
|
Purification and partial characterization of barley leucine aminopeptidase.
|
|
68985
|
Purification and partial characterization of barley leucine aminopeptidase.
|
|
68986
|
Purification and partial characterization of barley leucine aminopeptidase.
|
|
68987
|
Purification and partial characterization of barley leucine aminopeptidase.
|
|
68988
|
Purification and partial characterization of barley leucine aminopeptidase.
|
|
68989
|
Kininase activity in human platelets: cleavage of the Arg1-Pro2 bond of bradykinin by aminopeptidase P
|
|
68990
|
Kininase activity in human platelets: cleavage of the Arg1-Pro2 bond of bradykinin by aminopeptidase P
|
|
68991
|
A study of Bos taurus muscle specific enolase; biochemical characterization, homology modelling and ...
|
|
68992
|
Pantoate kinase and phosphopantothenate synthetase, two novel enzymes necessary for CoA biosynthesis in the ...
|
|
68993
|
Pantoate kinase and phosphopantothenate synthetase, two novel enzymes necessary for CoA biosynthesis in the ...
|
|
68994
|
Pantoate kinase and phosphopantothenate synthetase, two novel enzymes necessary for CoA biosynthesis in the ...
|
|
68995
|
Characterization of mammalian sedoheptulokinase and mechanism of formation of erythritol in sedoheptulokinase ...
|
|
68996
|
Characterization of mammalian sedoheptulokinase and mechanism of formation of erythritol in sedoheptulokinase ...
|
|
68997
|
Characterization of mammalian sedoheptulokinase and mechanism of formation of erythritol in sedoheptulokinase ...
|
|
68998
|
Characterization of mammalian sedoheptulokinase and mechanism of formation of erythritol in sedoheptulokinase ...
|
|
68999
|
Characterization of mammalian sedoheptulokinase and mechanism of formation of erythritol in sedoheptulokinase ...
|
|
69000
|
Catalysis, stereochemistry, and inhibition of ureidoglycolate lyase
|





 If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.
If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.