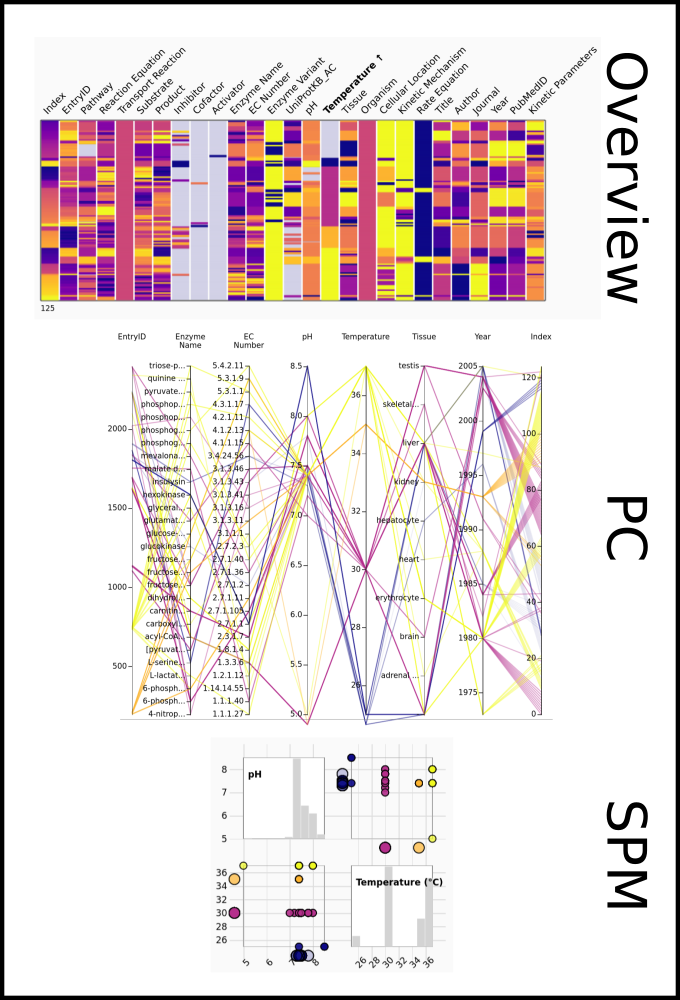
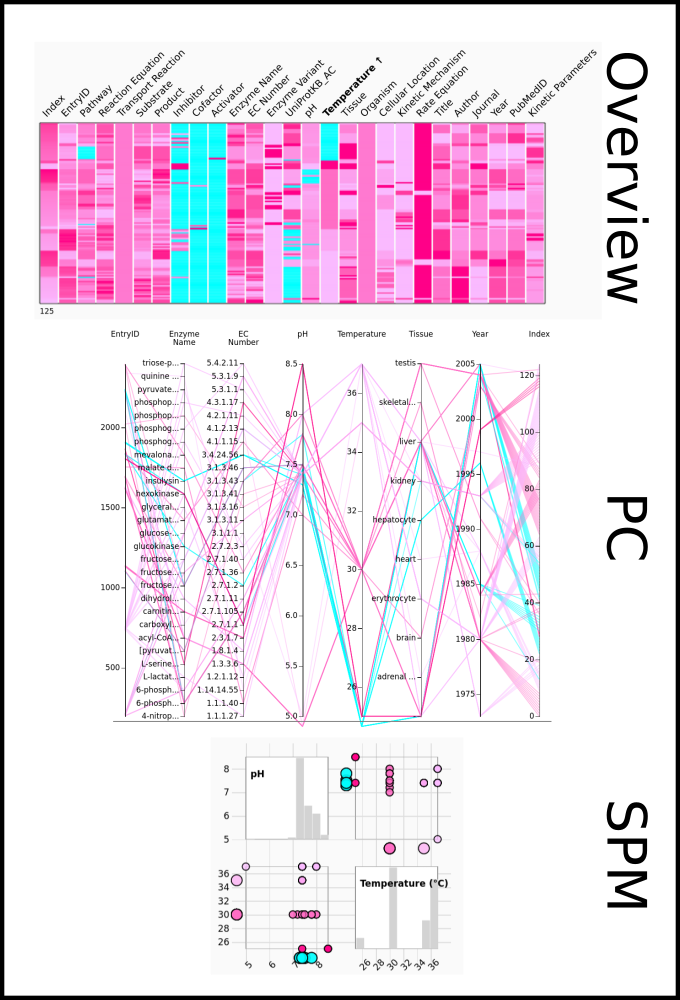
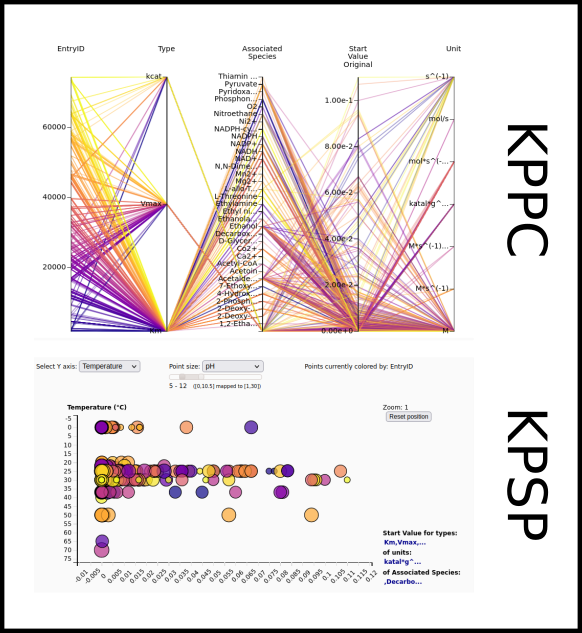
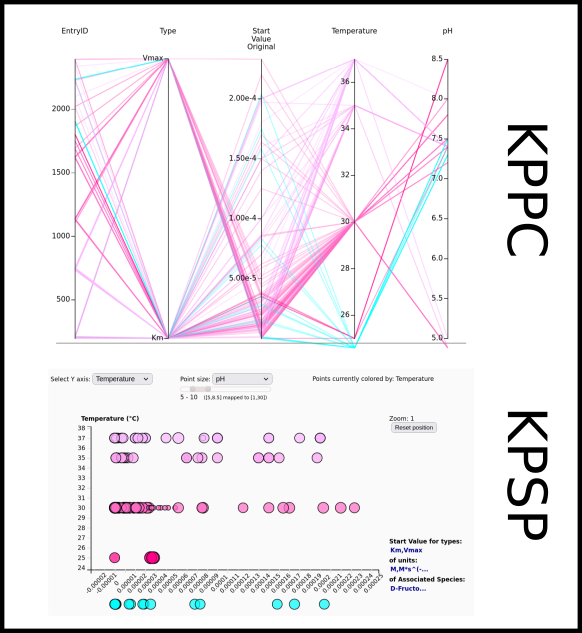
To improve SABIO-RK data interoperability, semantic markup was added to web pages as described and defined by
This structured information makes it easier to discover, collate and analyse our data.
|
65001
|
Molecular identification of a functional homologue of the mammalian fatty acid amide hydrolase in Arabidopsis ...
|
|
65002
|
Molecular identification of a functional homologue of the mammalian fatty acid amide hydrolase in Arabidopsis ...
|
|
65003
|
Molecular identification of a functional homologue of the mammalian fatty acid amide hydrolase in Arabidopsis ...
|
|
65004
|
Molecular identification of a functional homologue of the mammalian fatty acid amide hydrolase in Arabidopsis ...
|
|
65005
|
Molecular identification of a functional homologue of the mammalian fatty acid amide hydrolase in Arabidopsis ...
|
|
65006
|
Molecular identification of a functional homologue of the mammalian fatty acid amide hydrolase in Arabidopsis ...
|
|
65007
|
Molecular identification of a functional homologue of the mammalian fatty acid amide hydrolase in Arabidopsis ...
|
|
65008
|
Molecular identification of a functional homologue of the mammalian fatty acid amide hydrolase in Arabidopsis ...
|
|
65009
|
Molecular identification of a functional homologue of the mammalian fatty acid amide hydrolase in Arabidopsis ...
|
|
65010
|
Molecular identification of a functional homologue of the mammalian fatty acid amide hydrolase in Arabidopsis ...
|
|
65011
|
Biochemical Characterization and Complete Conversion of Coenzyme Specificity of Isocitrate Dehydrogenase from ...
|
|
65012
|
Biochemical Characterization and Complete Conversion of Coenzyme Specificity of Isocitrate Dehydrogenase from ...
|
|
65013
|
Biochemical Characterization and Complete Conversion of Coenzyme Specificity of Isocitrate Dehydrogenase from ...
|
|
65014
|
Biochemical Characterization and Complete Conversion of Coenzyme Specificity of Isocitrate Dehydrogenase from ...
|
|
65015
|
Biochemical Characterization and Complete Conversion of Coenzyme Specificity of Isocitrate Dehydrogenase from ...
|
|
65016
|
Biochemical Characterization and Complete Conversion of Coenzyme Specificity of Isocitrate Dehydrogenase from ...
|
|
65017
|
Biochemical Characterization and Complete Conversion of Coenzyme Specificity of Isocitrate Dehydrogenase from ...
|
|
65018
|
Biochemical Characterization and Complete Conversion of Coenzyme Specificity of Isocitrate Dehydrogenase from ...
|
|
65019
|
Biochemical Characterization and Complete Conversion of Coenzyme Specificity of Isocitrate Dehydrogenase from ...
|
|
65020
|
The gene mutated in l-2-hydroxyglutaric aciduria encodes l-2-hydroxyglutarate dehydrogenase
|
|
65021
|
The gene mutated in l-2-hydroxyglutaric aciduria encodes l-2-hydroxyglutarate dehydrogenase
|
|
65022
|
The gene mutated in l-2-hydroxyglutaric aciduria encodes l-2-hydroxyglutarate dehydrogenase
|
|
65023
|
The gene mutated in l-2-hydroxyglutaric aciduria encodes l-2-hydroxyglutarate dehydrogenase
|
|
65024
|
Enzymatic activity and partial purification of solanapyrone synthase: first enzyme catalyzing Diels-Alder ...
|
|
65025
|
Purification and characterization of sepiapterin reductase from rat erythrocytes.
|
|
65026
|
Purification and characterization of sepiapterin reductase from rat erythrocytes.
|
|
65027
|
Purification and characterization of sepiapterin reductase from rat erythrocytes.
|
|
65028
|
Structure based discovery of clomifene as a potent inhibitor of cancer-associated mutant IDH1
|
|
65029
|
Structure based discovery of clomifene as a potent inhibitor of cancer-associated mutant IDH1
|
|
65030
|
Structure based discovery of clomifene as a potent inhibitor of cancer-associated mutant IDH1
|
|
65031
|
Structure based discovery of clomifene as a potent inhibitor of cancer-associated mutant IDH1
|
|
65032
|
Structure based discovery of clomifene as a potent inhibitor of cancer-associated mutant IDH1
|
|
65033
|
Structure based discovery of clomifene as a potent inhibitor of cancer-associated mutant IDH1
|
|
65034
|
Structure based discovery of clomifene as a potent inhibitor of cancer-associated mutant IDH1
|
|
65035
|
Structure based discovery of clomifene as a potent inhibitor of cancer-associated mutant IDH1
|
|
65036
|
Structure based discovery of clomifene as a potent inhibitor of cancer-associated mutant IDH1
|
|
65037
|
Structure based discovery of clomifene as a potent inhibitor of cancer-associated mutant IDH1
|
|
65038
|
Structure based discovery of clomifene as a potent inhibitor of cancer-associated mutant IDH1
|
|
65039
|
Structure based discovery of clomifene as a potent inhibitor of cancer-associated mutant IDH1
|
|
65040
|
Distinct biochemical properties of human serine hydroxymethyltransferase compared with the Plasmodium enzyme: ...
|
|
65041
|
Distinct biochemical properties of human serine hydroxymethyltransferase compared with the Plasmodium enzyme: ...
|
|
65042
|
Distinct biochemical properties of human serine hydroxymethyltransferase compared with the Plasmodium enzyme: ...
|
|
65043
|
Distinct biochemical properties of human serine hydroxymethyltransferase compared with the Plasmodium enzyme: ...
|
|
65044
|
Distinct biochemical properties of human serine hydroxymethyltransferase compared with the Plasmodium enzyme: ...
|
|
65045
|
Distinct biochemical properties of human serine hydroxymethyltransferase compared with the Plasmodium enzyme: ...
|
|
65046
|
Distinct biochemical properties of human serine hydroxymethyltransferase compared with the Plasmodium enzyme: ...
|
|
65047
|
Distinct biochemical properties of human serine hydroxymethyltransferase compared with the Plasmodium enzyme: ...
|
|
65048
|
Distinct biochemical properties of human serine hydroxymethyltransferase compared with the Plasmodium enzyme: ...
|
|
65049
|
Optimized determination of gamma-glutamyltransferase by reaction-rate analysis.
|
|
65050
|
Optimized determination of gamma-glutamyltransferase by reaction-rate analysis.
|
|
65051
|
Isolation of gamma-glutamyltransferase from human liver, and comparison with the enzyme from human kidney.
|
|
65052
|
Isolation of gamma-glutamyltransferase from human liver, and comparison with the enzyme from human kidney.
|
|
65053
|
Isolation of gamma-glutamyltransferase from human liver, and comparison with the enzyme from human kidney.
|
|
65054
|
Isolation of gamma-glutamyltransferase from human liver, and comparison with the enzyme from human kidney.
|
|
65055
|
Isolation of gamma-glutamyltransferase from human liver, and comparison with the enzyme from human kidney.
|
|
65056
|
Isolation of gamma-glutamyltransferase from human liver, and comparison with the enzyme from human kidney.
|
|
65057
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65058
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65059
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65060
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65061
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65062
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65063
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65064
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65065
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65066
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65067
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65068
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65069
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65070
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65071
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65072
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65073
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65074
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65075
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65076
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65077
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65078
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65079
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65080
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65081
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65082
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65083
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65084
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65085
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65086
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65087
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65088
|
Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds
|
|
65089
|
Inhibition of Mutated Isocitrate Dehydrogenase 1 in Cancer
|
|
65090
|
Inhibition of Mutated Isocitrate Dehydrogenase 1 in Cancer
|
|
65091
|
Inhibition of Mutated Isocitrate Dehydrogenase 1 in Cancer
|
|
65092
|
Inhibition of Mutated Isocitrate Dehydrogenase 1 in Cancer
|
|
65093
|
Inhibition of Mutated Isocitrate Dehydrogenase 1 in Cancer
|
|
65094
|
Inhibition of Mutated Isocitrate Dehydrogenase 1 in Cancer
|
|
65095
|
Inhibition of Mutated Isocitrate Dehydrogenase 1 in Cancer
|
|
65096
|
Inhibition of Mutated Isocitrate Dehydrogenase 1 in Cancer
|
|
65097
|
Inhibition of Mutated Isocitrate Dehydrogenase 1 in Cancer
|
|
65098
|
Inhibition of Mutated Isocitrate Dehydrogenase 1 in Cancer
|
|
65099
|
Inhibition of Mutated Isocitrate Dehydrogenase 1 in Cancer
|
|
65100
|
Inhibition of Mutated Isocitrate Dehydrogenase 1 in Cancer
|
|
65101
|
Inhibition of Mutated Isocitrate Dehydrogenase 1 in Cancer
|
|
65102
|
Inhibition of Mutated Isocitrate Dehydrogenase 1 in Cancer
|
|
65103
|
Selective inhibition of mutant isocitrate dehydrogenase 1 (IDH1) via disruption of a metal binding network by ...
|
|
65104
|
Selective inhibition of mutant isocitrate dehydrogenase 1 (IDH1) via disruption of a metal binding network by ...
|
|
65105
|
Selective inhibition of mutant isocitrate dehydrogenase 1 (IDH1) via disruption of a metal binding network by ...
|
|
65106
|
Selective inhibition of mutant isocitrate dehydrogenase 1 (IDH1) via disruption of a metal binding network by ...
|
|
65107
|
Selective inhibition of mutant isocitrate dehydrogenase 1 (IDH1) via disruption of a metal binding network by ...
|
|
65108
|
Selective inhibition of mutant isocitrate dehydrogenase 1 (IDH1) via disruption of a metal binding network by ...
|
|
65109
|
Selective inhibition of mutant isocitrate dehydrogenase 1 (IDH1) via disruption of a metal binding network by ...
|
|
65110
|
Selective inhibition of mutant isocitrate dehydrogenase 1 (IDH1) via disruption of a metal binding network by ...
|
|
65111
|
Selective inhibition of mutant isocitrate dehydrogenase 1 (IDH1) via disruption of a metal binding network by ...
|
|
65112
|
Selective inhibition of mutant isocitrate dehydrogenase 1 (IDH1) via disruption of a metal binding network by ...
|
|
65113
|
Selective inhibition of mutant isocitrate dehydrogenase 1 (IDH1) via disruption of a metal binding network by ...
|
|
65114
|
Selective inhibition of mutant isocitrate dehydrogenase 1 (IDH1) via disruption of a metal binding network by ...
|
|
65115
|
Selective inhibition of mutant isocitrate dehydrogenase 1 (IDH1) via disruption of a metal binding network by ...
|
|
65116
|
Selective inhibition of mutant isocitrate dehydrogenase 1 (IDH1) via disruption of a metal binding network by ...
|
|
65117
|
Selective inhibition of mutant isocitrate dehydrogenase 1 (IDH1) via disruption of a metal binding network by ...
|
|
65118
|
Selective inhibition of mutant isocitrate dehydrogenase 1 (IDH1) via disruption of a metal binding network by ...
|
|
65119
|
Selective inhibition of mutant isocitrate dehydrogenase 1 (IDH1) via disruption of a metal binding network by ...
|
|
65120
|
Selective inhibition of mutant isocitrate dehydrogenase 1 (IDH1) via disruption of a metal binding network by ...
|
|
65121
|
Selective inhibition of mutant isocitrate dehydrogenase 1 (IDH1) via disruption of a metal binding network by ...
|
|
65122
|
Selective inhibition of mutant isocitrate dehydrogenase 1 (IDH1) via disruption of a metal binding network by ...
|
|
65123
|
Selective inhibition of mutant isocitrate dehydrogenase 1 (IDH1) via disruption of a metal binding network by ...
|
|
65124
|
Extremely stable indole-3-glycerol-phosphate synthase from hyperthermophilic archaeon Pyrococcus furiosus
|
|
65125
|
Metabolic interactions of rosmarinic acid with human cytochrome P450 monooxygenases and uridine diphosphate ...
|
|
65126
|
Metabolic interactions of rosmarinic acid with human cytochrome P450 monooxygenases and uridine diphosphate ...
|
|
65127
|
Metabolic interactions of rosmarinic acid with human cytochrome P450 monooxygenases and uridine diphosphate ...
|
|
65128
|
Metabolic interactions of rosmarinic acid with human cytochrome P450 monooxygenases and uridine diphosphate ...
|
|
65129
|
Metabolic interactions of rosmarinic acid with human cytochrome P450 monooxygenases and uridine diphosphate ...
|
|
65130
|
Metabolic interactions of rosmarinic acid with human cytochrome P450 monooxygenases and uridine diphosphate ...
|
|
65131
|
Metabolic interactions of rosmarinic acid with human cytochrome P450 monooxygenases and uridine diphosphate ...
|
|
65132
|
Metabolic interactions of rosmarinic acid with human cytochrome P450 monooxygenases and uridine diphosphate ...
|
|
65133
|
Single Amino Acid Change in 6-Phosphogluconate Dehydrogenase from Synechocystis Conveys Higher Affinity for ...
|
|
65134
|
Single Amino Acid Change in 6-Phosphogluconate Dehydrogenase from Synechocystis Conveys Higher Affinity for ...
|
|
65135
|
Single Amino Acid Change in 6-Phosphogluconate Dehydrogenase from Synechocystis Conveys Higher Affinity for ...
|
|
65136
|
Single Amino Acid Change in 6-Phosphogluconate Dehydrogenase from Synechocystis Conveys Higher Affinity for ...
|
|
65137
|
Single Amino Acid Change in 6-Phosphogluconate Dehydrogenase from Synechocystis Conveys Higher Affinity for ...
|
|
65138
|
Single Amino Acid Change in 6-Phosphogluconate Dehydrogenase from Synechocystis Conveys Higher Affinity for ...
|
|
65139
|
Single Amino Acid Change in 6-Phosphogluconate Dehydrogenase from Synechocystis Conveys Higher Affinity for ...
|
|
65140
|
Single Amino Acid Change in 6-Phosphogluconate Dehydrogenase from Synechocystis Conveys Higher Affinity for ...
|
|
65141
|
Single Amino Acid Change in 6-Phosphogluconate Dehydrogenase from Synechocystis Conveys Higher Affinity for ...
|
|
65142
|
Single Amino Acid Change in 6-Phosphogluconate Dehydrogenase from Synechocystis Conveys Higher Affinity for ...
|
|
65143
|
Single Amino Acid Change in 6-Phosphogluconate Dehydrogenase from Synechocystis Conveys Higher Affinity for ...
|
|
65144
|
Single Amino Acid Change in 6-Phosphogluconate Dehydrogenase from Synechocystis Conveys Higher Affinity for ...
|
|
65145
|
Single Amino Acid Change in 6-Phosphogluconate Dehydrogenase from Synechocystis Conveys Higher Affinity for ...
|
|
65146
|
Single Amino Acid Change in 6-Phosphogluconate Dehydrogenase from Synechocystis Conveys Higher Affinity for ...
|
|
65147
|
Single Amino Acid Change in 6-Phosphogluconate Dehydrogenase from Synechocystis Conveys Higher Affinity for ...
|
|
65148
|
Single Amino Acid Change in 6-Phosphogluconate Dehydrogenase from Synechocystis Conveys Higher Affinity for ...
|
|
65149
|
Characterization of biochemical properties of an apurinic/apyrimidinic endonuclease from Helicobacter pylori.
|
|
65150
|
Characterization of biochemical properties of an apurinic/apyrimidinic endonuclease from Helicobacter pylori.
|
|
65151
|
Characterization of biochemical properties of an apurinic/apyrimidinic endonuclease from Helicobacter pylori.
|
|
65152
|
Characterization of biochemical properties of an apurinic/apyrimidinic endonuclease from Helicobacter pylori.
|
|
65153
|
Lipocalin-type prostaglandin D synthase (beta-trace) is a newly recognized type of retinoid transporter
|
|
65154
|
Lipocalin-type prostaglandin D synthase (beta-trace) is a newly recognized type of retinoid transporter
|
|
65155
|
Lipocalin-type prostaglandin D synthase (beta-trace) is a newly recognized type of retinoid transporter
|
|
65156
|
Lipocalin-type prostaglandin D synthase (beta-trace) is a newly recognized type of retinoid transporter
|
|
65157
|
Lipocalin-type prostaglandin D synthase (beta-trace) is a newly recognized type of retinoid transporter
|
|
65158
|
Lipocalin-type prostaglandin D synthase (beta-trace) is a newly recognized type of retinoid transporter
|
|
65159
|
Human phosphoglycerate dehydrogenase produces the oncometabolite D-2-hydroxyglutarate
|
|
65160
|
Human phosphoglycerate dehydrogenase produces the oncometabolite D-2-hydroxyglutarate
|
|
65161
|
Human phosphoglycerate dehydrogenase produces the oncometabolite D-2-hydroxyglutarate
|
|
65162
|
Human phosphoglycerate dehydrogenase produces the oncometabolite D-2-hydroxyglutarate
|
|
65163
|
Human phosphoglycerate dehydrogenase produces the oncometabolite D-2-hydroxyglutarate
|
|
65164
|
Human phosphoglycerate dehydrogenase produces the oncometabolite D-2-hydroxyglutarate
|
|
65165
|
Molecular characterization of cytosolic cysteine synthase in Mimosa pudica
|
|
65166
|
Molecular characterization of cytosolic cysteine synthase in Mimosa pudica
|
|
65167
|
Purification and characterization of novel bi-functional GH3 family β-xylosidase/β-glucosidase from ...
|
|
65168
|
Purification and characterization of a hydroxynitrile lyase from Amygdalus pedunculata Pall
|
|
65169
|
B-factor-saturation mutagenesis as a strategy to increase the thermostability of α-L-rhamnosidase from ...
|
|
65170
|
B-factor-saturation mutagenesis as a strategy to increase the thermostability of α-L-rhamnosidase from ...
|
|
65171
|
B-factor-saturation mutagenesis as a strategy to increase the thermostability of α-L-rhamnosidase from ...
|
|
65172
|
B-factor-saturation mutagenesis as a strategy to increase the thermostability of α-L-rhamnosidase from ...
|
|
65173
|
B-factor-saturation mutagenesis as a strategy to increase the thermostability of α-L-rhamnosidase from ...
|
|
65174
|
B-factor-saturation mutagenesis as a strategy to increase the thermostability of α-L-rhamnosidase from ...
|
|
65175
|
Receptor protein of Lysinibacillus sphaericus mosquito-larvicidal toxin displays amylomaltase activity
|
|
65176
|
Receptor protein of Lysinibacillus sphaericus mosquito-larvicidal toxin displays amylomaltase activity
|
|
65177
|
Receptor protein of Lysinibacillus sphaericus mosquito-larvicidal toxin displays amylomaltase activity
|
|
65178
|
The yeast inositol monophosphatase is a lithium- and sodium-sensitive enzyme encoded by a non-essential gene ...
|
|
65179
|
The yeast inositol monophosphatase is a lithium- and sodium-sensitive enzyme encoded by a non-essential gene ...
|
|
65180
|
The yeast inositol monophosphatase is a lithium- and sodium-sensitive enzyme encoded by a non-essential gene ...
|
|
65181
|
The yeast inositol monophosphatase is a lithium- and sodium-sensitive enzyme encoded by a non-essential gene ...
|
|
65182
|
The yeast inositol monophosphatase is a lithium- and sodium-sensitive enzyme encoded by a non-essential gene ...
|
|
65183
|
The yeast inositol monophosphatase is a lithium- and sodium-sensitive enzyme encoded by a non-essential gene ...
|
|
65184
|
Identification and characterization of a sac domain-containing phosphoinositide 5-phosphatase
|
|
65185
|
Purification, molecular cloning, and sequence analysis of sucrose-6F-phosphate phosphohydrolase from plants
|
|
65186
|
Purification, molecular cloning, and sequence analysis of sucrose-6F-phosphate phosphohydrolase from plants
|
|
65187
|
Purification, molecular cloning, and sequence analysis of sucrose-6F-phosphate phosphohydrolase from plants
|
|
65188
|
Characterization of kinetics of human cytochrome P450s involved in bioactivation of flucloxacillin: inhibition ...
|
|
65189
|
Characterization of kinetics of human cytochrome P450s involved in bioactivation of flucloxacillin: inhibition ...
|
|
65190
|
Characterization of kinetics of human cytochrome P450s involved in bioactivation of flucloxacillin: inhibition ...
|
|
65191
|
Characterization of kinetics of human cytochrome P450s involved in bioactivation of flucloxacillin: inhibition ...
|
|
65192
|
Characterization of kinetics of human cytochrome P450s involved in bioactivation of flucloxacillin: inhibition ...
|
|
65193
|
Characterization of kinetics of human cytochrome P450s involved in bioactivation of flucloxacillin: inhibition ...
|
|
65194
|
Characterization of kinetics of human cytochrome P450s involved in bioactivation of flucloxacillin: inhibition ...
|
|
65195
|
Characterization of kinetics of human cytochrome P450s involved in bioactivation of flucloxacillin: inhibition ...
|
|
65196
|
Kinetics of manganese lipoxygenase with a catalytic mononuclear redox center
|
|
65197
|
Kinetics of manganese lipoxygenase with a catalytic mononuclear redox center
|
|
65198
|
Kinetics of manganese lipoxygenase with a catalytic mononuclear redox center
|
|
65199
|
Kinetics of manganese lipoxygenase with a catalytic mononuclear redox center
|
|
65200
|
Kinetics of manganese lipoxygenase with a catalytic mononuclear redox center
|
|
65201
|
Purification and characterization of linoleate 8-dioxygenase from the fungus Gaeumannomyces graminis as a ...
|
|
65202
|
Purification and characterization of linoleate 8-dioxygenase from the fungus Gaeumannomyces graminis as a ...
|
|
65203
|
Purification and characterization of linoleate 8-dioxygenase from the fungus Gaeumannomyces graminis as a ...
|
|
65204
|
Purification and characterization of linoleate 8-dioxygenase from the fungus Gaeumannomyces graminis as a ...
|
|
65205
|
Identification and reconstitution of the yeast mitochondrial transporter for thiamine pyrophosphate
|
|
65206
|
Identification and reconstitution of the yeast mitochondrial transporter for thiamine pyrophosphate
|
|
65207
|
Identification and reconstitution of the yeast mitochondrial transporter for thiamine pyrophosphate
|
|
65208
|
Identification and reconstitution of the yeast mitochondrial transporter for thiamine pyrophosphate
|
|
65209
|
Identification and reconstitution of the yeast mitochondrial transporter for thiamine pyrophosphate
|
|
65210
|
Identification and reconstitution of the yeast mitochondrial transporter for thiamine pyrophosphate
|
|
65211
|
High-activity production of xylanase by Pichia stipitis: Purification, characterization, kinetic evaluation ...
|
|
65212
|
A rapid and sensitive colorimetric assay for the determination of adenosine kinase activity
|
|
65213
|
Characterization of the enzymatic activity of human kallikrein 6: Autoactivation, substrate specificity, and ...
|
|
65214
|
Characterization of the enzymatic activity of human kallikrein 6: Autoactivation, substrate specificity, and ...
|
|
65215
|
Characterization of the enzymatic activity of human kallikrein 6: Autoactivation, substrate specificity, and ...
|
|
65216
|
Characterization of the enzymatic activity of human kallikrein 6: Autoactivation, substrate specificity, and ...
|
|
65217
|
Characterization of the enzymatic activity of human kallikrein 6: Autoactivation, substrate specificity, and ...
|
|
65218
|
Characterization of the enzymatic activity of human kallikrein 6: Autoactivation, substrate specificity, and ...
|
|
65219
|
Characterization of the enzymatic activity of human kallikrein 6: Autoactivation, substrate specificity, and ...
|
|
65220
|
Structural and mutational analysis of substrate complexation by anthranilate phosphoribosyltransferase from ...
|
|
65221
|
Structural and mutational analysis of substrate complexation by anthranilate phosphoribosyltransferase from ...
|
|
65222
|
Structural and mutational analysis of substrate complexation by anthranilate phosphoribosyltransferase from ...
|
|
65223
|
Structural and mutational analysis of substrate complexation by anthranilate phosphoribosyltransferase from ...
|
|
65224
|
Structural and mutational analysis of substrate complexation by anthranilate phosphoribosyltransferase from ...
|
|
65225
|
Structural and mutational analysis of substrate complexation by anthranilate phosphoribosyltransferase from ...
|
|
65226
|
Structural and mutational analysis of substrate complexation by anthranilate phosphoribosyltransferase from ...
|
|
65227
|
Structural and mutational analysis of substrate complexation by anthranilate phosphoribosyltransferase from ...
|
|
65228
|
Structural and mutational analysis of substrate complexation by anthranilate phosphoribosyltransferase from ...
|
|
65229
|
Hint2, a mitochondrial apoptotic sensitizer down-regulated in hepatocellular carcinoma
|
|
65230
|
Evaluation of the activity of β-glucosidase immobilized on polydimethylsiloxane (PDMS) with a microfluidic ...
|
|
65231
|
Evaluation of the activity of β-glucosidase immobilized on polydimethylsiloxane (PDMS) with a microfluidic ...
|
|
65232
|
Identity of SMCT1 (SLC5A8) as a neuron-specific Na+-coupled transporter for active uptake of L-lactate and ...
|
|
65233
|
Identity of SMCT1 (SLC5A8) as a neuron-specific Na+-coupled transporter for active uptake of L-lactate and ...
|
|
65234
|
Identity of SMCT1 (SLC5A8) as a neuron-specific Na+-coupled transporter for active uptake of L-lactate and ...
|
|
65235
|
Identity of SMCT1 (SLC5A8) as a neuron-specific Na+-coupled transporter for active uptake of L-lactate and ...
|
|
65236
|
Identity of SMCT1 (SLC5A8) as a neuron-specific Na+-coupled transporter for active uptake of L-lactate and ...
|
|
65237
|
Identity of SMCT1 (SLC5A8) as a neuron-specific Na+-coupled transporter for active uptake of L-lactate and ...
|
|
65238
|
Identity of SMCT1 (SLC5A8) as a neuron-specific Na+-coupled transporter for active uptake of L-lactate and ...
|
|
65239
|
Identity of SMCT1 (SLC5A8) as a neuron-specific Na+-coupled transporter for active uptake of L-lactate and ...
|
|
65240
|
Structural and Functional Analysis of UGT92G6 Suggests an Evolutionary Link Between Mono- and Disaccharide ...
|
|
65241
|
Structural and Functional Analysis of UGT92G6 Suggests an Evolutionary Link Between Mono- and Disaccharide ...
|
|
65242
|
Structural and Functional Analysis of UGT92G6 Suggests an Evolutionary Link Between Mono- and Disaccharide ...
|
|
65243
|
The effect of alpha-solanine on the activity, gene expression, and kinetics of arylamine N-acetyltransferase ...
|
|
65244
|
The effect of alpha-solanine on the activity, gene expression, and kinetics of arylamine N-acetyltransferase ...
|
|
65245
|
The effect of alpha-solanine on the activity, gene expression, and kinetics of arylamine N-acetyltransferase ...
|
|
65246
|
The effect of alpha-solanine on the activity, gene expression, and kinetics of arylamine N-acetyltransferase ...
|
|
65247
|
Zinc ion acts as a cofactor for serine/threonine kinase MST3 and has a distinct role in autophosphorylation of ...
|
|
65248
|
Zinc ion acts as a cofactor for serine/threonine kinase MST3 and has a distinct role in autophosphorylation of ...
|
|
65249
|
Zinc ion acts as a cofactor for serine/threonine kinase MST3 and has a distinct role in autophosphorylation of ...
|
|
65250
|
Zinc ion acts as a cofactor for serine/threonine kinase MST3 and has a distinct role in autophosphorylation of ...
|
|
65251
|
Cep-1347 (KT7515), a semisynthetic inhibitor of the mixed lineage kinase family
|
|
65252
|
Cep-1347 (KT7515), a semisynthetic inhibitor of the mixed lineage kinase family
|
|
65253
|
Cep-1347 (KT7515), a semisynthetic inhibitor of the mixed lineage kinase family
|
|
65254
|
Cep-1347 (KT7515), a semisynthetic inhibitor of the mixed lineage kinase family
|
|
65255
|
Cep-1347 (KT7515), a semisynthetic inhibitor of the mixed lineage kinase family
|
|
65256
|
Cep-1347 (KT7515), a semisynthetic inhibitor of the mixed lineage kinase family
|
|
65257
|
Cep-1347 (KT7515), a semisynthetic inhibitor of the mixed lineage kinase family
|
|
65258
|
Cep-1347 (KT7515), a semisynthetic inhibitor of the mixed lineage kinase family
|
|
65259
|
Cep-1347 (KT7515), a semisynthetic inhibitor of the mixed lineage kinase family
|
|
65260
|
Molecular and biochemical characterization of two brassinosteroid sulfotransferases from Arabidopsis, AtST4a ...
|
|
65261
|
Molecular and biochemical characterization of two brassinosteroid sulfotransferases from Arabidopsis, AtST4a ...
|
|
65262
|
Molecular and biochemical characterization of two brassinosteroid sulfotransferases from Arabidopsis, AtST4a ...
|
|
65263
|
Molecular and biochemical characterization of two brassinosteroid sulfotransferases from Arabidopsis, AtST4a ...
|
|
65264
|
Molecular and biochemical characterization of two brassinosteroid sulfotransferases from Arabidopsis, AtST4a ...
|
|
65265
|
Molecular and biochemical characterization of two brassinosteroid sulfotransferases from Arabidopsis, AtST4a ...
|
|
65266
|
Molecular and biochemical characterization of two brassinosteroid sulfotransferases from Arabidopsis, AtST4a ...
|
|
65267
|
Molecular and biochemical characterization of two brassinosteroid sulfotransferases from Arabidopsis, AtST4a ...
|
|
65268
|
Molecular and biochemical characterization of two brassinosteroid sulfotransferases from Arabidopsis, AtST4a ...
|
|
65269
|
Molecular and biochemical characterization of two brassinosteroid sulfotransferases from Arabidopsis, AtST4a ...
|
|
65270
|
Molecular and biochemical characterization of two brassinosteroid sulfotransferases from Arabidopsis, AtST4a ...
|
|
65271
|
Molecular and biochemical characterization of two brassinosteroid sulfotransferases from Arabidopsis, AtST4a ...
|
|
65272
|
Molecular and biochemical characterization of two brassinosteroid sulfotransferases from Arabidopsis, AtST4a ...
|
|
65273
|
Molecular and biochemical characterization of two brassinosteroid sulfotransferases from Arabidopsis, AtST4a ...
|
|
65274
|
Cutting Edge: IL-1 receptor-associated kinase 4 structures reveal novel features and multiple conformations
|
|
65275
|
Cutting Edge: IL-1 receptor-associated kinase 4 structures reveal novel features and multiple conformations
|
|
65276
|
Cutting Edge: IL-1 receptor-associated kinase 4 structures reveal novel features and multiple conformations
|
|
65277
|
PRKX, a phylogenetically and functionally distinct cAMP-dependent protein kinase, activates renal epithelial ...
|
|
65278
|
PRKX, a phylogenetically and functionally distinct cAMP-dependent protein kinase, activates renal epithelial ...
|
|
65279
|
Biochemical characterization of the kinase domain of the rice disease resistance receptor-like kinase XA21
|
|
65280
|
A protein kinase family in Arabidopsis phosphorylates chloroplast precursor proteins
|
|
65281
|
A protein kinase family in Arabidopsis phosphorylates chloroplast precursor proteins
|
|
65282
|
A protein kinase family in Arabidopsis phosphorylates chloroplast precursor proteins
|
|
65283
|
Calcium/calmodulin up-regulates a cytoplasmic receptor-like kinase in plants
|
|
65284
|
Calcium/calmodulin up-regulates a cytoplasmic receptor-like kinase in plants
|
|
65285
|
Calcium/calmodulin up-regulates a cytoplasmic receptor-like kinase in plants
|
|
65286
|
Novel substrates of Mycobacterium tuberculosis PknH Ser/Thr kinase
|
|
65287
|
Novel substrates of Mycobacterium tuberculosis PknH Ser/Thr kinase
|
|
65288
|
Novel substrates of Mycobacterium tuberculosis PknH Ser/Thr kinase
|
|
65289
|
TTBK2 kinase substrate specificity and the impact of spinocerebellar-ataxia-causing mutations on expression, ...
|
|
65290
|
TTBK2 kinase substrate specificity and the impact of spinocerebellar-ataxia-causing mutations on expression, ...
|
|
65291
|
TTBK2 kinase substrate specificity and the impact of spinocerebellar-ataxia-causing mutations on expression, ...
|
|
65292
|
TTBK2 kinase substrate specificity and the impact of spinocerebellar-ataxia-causing mutations on expression, ...
|
|
65293
|
TTBK2 kinase substrate specificity and the impact of spinocerebellar-ataxia-causing mutations on expression, ...
|
|
65294
|
TTBK2 kinase substrate specificity and the impact of spinocerebellar-ataxia-causing mutations on expression, ...
|
|
65295
|
TTBK2 kinase substrate specificity and the impact of spinocerebellar-ataxia-causing mutations on expression, ...
|
|
65296
|
TTBK2 kinase substrate specificity and the impact of spinocerebellar-ataxia-causing mutations on expression, ...
|
|
65297
|
TTBK2 kinase substrate specificity and the impact of spinocerebellar-ataxia-causing mutations on expression, ...
|
|
65298
|
TTBK2 kinase substrate specificity and the impact of spinocerebellar-ataxia-causing mutations on expression, ...
|
|
65299
|
TTBK2 kinase substrate specificity and the impact of spinocerebellar-ataxia-causing mutations on expression, ...
|
|
65300
|
TTBK2 kinase substrate specificity and the impact of spinocerebellar-ataxia-causing mutations on expression, ...
|
|
65301
|
TTBK2 kinase substrate specificity and the impact of spinocerebellar-ataxia-causing mutations on expression, ...
|
|
65302
|
TTBK2 kinase substrate specificity and the impact of spinocerebellar-ataxia-causing mutations on expression, ...
|
|
65303
|
TTBK2 kinase substrate specificity and the impact of spinocerebellar-ataxia-causing mutations on expression, ...
|
|
65304
|
TTBK2 kinase substrate specificity and the impact of spinocerebellar-ataxia-causing mutations on expression, ...
|
|
65305
|
TTBK2 kinase substrate specificity and the impact of spinocerebellar-ataxia-causing mutations on expression, ...
|
|
65306
|
TTBK2 kinase substrate specificity and the impact of spinocerebellar-ataxia-causing mutations on expression, ...
|
|
65307
|
TTBK2 kinase substrate specificity and the impact of spinocerebellar-ataxia-causing mutations on expression, ...
|
|
65308
|
TTBK2 kinase substrate specificity and the impact of spinocerebellar-ataxia-causing mutations on expression, ...
|
|
65309
|
TTBK2 kinase substrate specificity and the impact of spinocerebellar-ataxia-causing mutations on expression, ...
|
|
65310
|
Phosphorylation of calcineurin B-like (CBL) calcium sensor proteins by their CBL-interacting protein kinases ...
|
|
65311
|
Phosphorylation of calcineurin B-like (CBL) calcium sensor proteins by their CBL-interacting protein kinases ...
|
|
65312
|
Phosphorylation of calcineurin B-like (CBL) calcium sensor proteins by their CBL-interacting protein kinases ...
|
|
65313
|
Phosphorylation of calcineurin B-like (CBL) calcium sensor proteins by their CBL-interacting protein kinases ...
|
|
65314
|
Phosphorylation of calcineurin B-like (CBL) calcium sensor proteins by their CBL-interacting protein kinases ...
|
|
65315
|
Phosphorylation of calcineurin B-like (CBL) calcium sensor proteins by their CBL-interacting protein kinases ...
|
|
65316
|
Ca(2+)/Calmodulin-dependent protein kinase cascade in Caenorhabditis elegans. Implication in transcriptional ...
|
|
65317
|
Ca(2+)/Calmodulin-dependent protein kinase cascade in Caenorhabditis elegans. Implication in transcriptional ...
|
|
65318
|
Ca(2+)/Calmodulin-dependent protein kinase cascade in Caenorhabditis elegans. Implication in transcriptional ...
|
|
65319
|
Ca(2+)/Calmodulin-dependent protein kinase cascade in Caenorhabditis elegans. Implication in transcriptional ...
|
|
65320
|
Phosphorylation of GRK1 and GRK7 by cAMP-dependent protein kinase attenuates their enzymatic activities
|
|
65321
|
Phosphorylation of GRK1 and GRK7 by cAMP-dependent protein kinase attenuates their enzymatic activities
|
|
65322
|
Phosphorylation of GRK1 and GRK7 by cAMP-dependent protein kinase attenuates their enzymatic activities
|
|
65323
|
Phosphorylation of GRK1 and GRK7 by cAMP-dependent protein kinase attenuates their enzymatic activities
|
|
65324
|
GRK1 and GRK7: unique cellular distribution and widely different activities of opsin phosphorylation in the ...
|
|
65325
|
GRK1 and GRK7: unique cellular distribution and widely different activities of opsin phosphorylation in the ...
|
|
65326
|
GRK1 and GRK7: unique cellular distribution and widely different activities of opsin phosphorylation in the ...
|
|
65327
|
GRK1 and GRK7: unique cellular distribution and widely different activities of opsin phosphorylation in the ...
|
|
65328
|
Key residues in Mycobacterium tuberculosis protein kinase G play a role in regulating kinase activity and ...
|
|
65329
|
Key residues in Mycobacterium tuberculosis protein kinase G play a role in regulating kinase activity and ...
|
|
65330
|
Key residues in Mycobacterium tuberculosis protein kinase G play a role in regulating kinase activity and ...
|
|
65331
|
Crystal structure of the Golgi casein kinase
|
|
65332
|
Crystal structure of the Golgi casein kinase
|
|
65333
|
Crystal structure of the Golgi casein kinase
|
|
65334
|
Crystal structure of the Golgi casein kinase
|
|
65335
|
Identification of cardiac-specific myosin light chain kinase
|
|
65336
|
Identification of cardiac-specific myosin light chain kinase
|
|
65337
|
Identification of cardiac-specific myosin light chain kinase
|
|
65338
|
The tricarboxylic acid cycle of Helicobacter pylori
|
|
65339
|
The tricarboxylic acid cycle of Helicobacter pylori
|
|
65340
|
The tricarboxylic acid cycle of Helicobacter pylori
|
|
65341
|
The tricarboxylic acid cycle of Helicobacter pylori
|
|
65342
|
The tricarboxylic acid cycle of Helicobacter pylori
|
|
65343
|
The tricarboxylic acid cycle of Helicobacter pylori
|
|
65344
|
The tricarboxylic acid cycle of Helicobacter pylori
|
|
65345
|
The tricarboxylic acid cycle of Helicobacter pylori
|
|
65346
|
The tricarboxylic acid cycle of Helicobacter pylori
|
|
65347
|
The tricarboxylic acid cycle of Helicobacter pylori
|
|
65348
|
The tricarboxylic acid cycle of Helicobacter pylori
|
|
65349
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65350
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65351
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65352
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65353
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65354
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65355
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65356
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65357
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65358
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65359
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65360
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65361
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65362
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65363
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65364
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65365
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65366
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65367
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65368
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65369
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65370
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65371
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65372
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65373
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65374
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65375
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65376
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65377
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65378
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65379
|
New IDH1 mutant inhibitors for treatment of acute myeloid leukemia
|
|
65380
|
Biochemical, cellular, and biophysical characterization of a potent inhibitor of mutant isocitrate ...
|
|
65381
|
Biochemical, cellular, and biophysical characterization of a potent inhibitor of mutant isocitrate ...
|
|
65382
|
Biochemical, cellular, and biophysical characterization of a potent inhibitor of mutant isocitrate ...
|
|
65383
|
Biochemical, cellular, and biophysical characterization of a potent inhibitor of mutant isocitrate ...
|
|
65384
|
Biochemical, cellular, and biophysical characterization of a potent inhibitor of mutant isocitrate ...
|
|
65385
|
Biochemical, cellular, and biophysical characterization of a potent inhibitor of mutant isocitrate ...
|
|
65386
|
Biochemical, cellular, and biophysical characterization of a potent inhibitor of mutant isocitrate ...
|
|
65387
|
Biochemical, cellular, and biophysical characterization of a potent inhibitor of mutant isocitrate ...
|
|
65388
|
Biochemical, cellular, and biophysical characterization of a potent inhibitor of mutant isocitrate ...
|
|
65389
|
Biochemical, cellular, and biophysical characterization of a potent inhibitor of mutant isocitrate ...
|
|
65390
|
Biochemical, cellular, and biophysical characterization of a potent inhibitor of mutant isocitrate ...
|
|
65391
|
Biochemical, cellular, and biophysical characterization of a potent inhibitor of mutant isocitrate ...
|
|
65392
|
Biochemical, cellular, and biophysical characterization of a potent inhibitor of mutant isocitrate ...
|
|
65393
|
Biochemical, cellular, and biophysical characterization of a potent inhibitor of mutant isocitrate ...
|
|
65394
|
Biochemical, cellular, and biophysical characterization of a potent inhibitor of mutant isocitrate ...
|
|
65395
|
Biochemical, cellular, and biophysical characterization of a potent inhibitor of mutant isocitrate ...
|
|
65396
|
Biochemical, cellular, and biophysical characterization of a potent inhibitor of mutant isocitrate ...
|
|
65397
|
Biochemical, cellular, and biophysical characterization of a potent inhibitor of mutant isocitrate ...
|
|
65398
|
Biochemical, cellular, and biophysical characterization of a potent inhibitor of mutant isocitrate ...
|
|
65399
|
Characterization of the physicochemical and catalytic properties of human heart NADP-dependent isocitrate ...
|
|
65400
|
Characterization of the physicochemical and catalytic properties of human heart NADP-dependent isocitrate ...
|
|
65401
|
Characterization of the physicochemical and catalytic properties of human heart NADP-dependent isocitrate ...
|
|
65402
|
Characterization of the physicochemical and catalytic properties of human heart NADP-dependent isocitrate ...
|
|
65403
|
Characterization of the physicochemical and catalytic properties of human heart NADP-dependent isocitrate ...
|
|
65404
|
Characterization of the physicochemical and catalytic properties of human heart NADP-dependent isocitrate ...
|
|
65405
|
Characterization of the physicochemical and catalytic properties of human heart NADP-dependent isocitrate ...
|
|
65406
|
Characterization of the physicochemical and catalytic properties of human heart NADP-dependent isocitrate ...
|
|
65407
|
Characterization of the physicochemical and catalytic properties of human heart NADP-dependent isocitrate ...
|
|
65408
|
Characterization of the physicochemical and catalytic properties of human heart NADP-dependent isocitrate ...
|
|
65409
|
Characterization of the physicochemical and catalytic properties of human heart NADP-dependent isocitrate ...
|
|
65410
|
Characterization of the physicochemical and catalytic properties of human heart NADP-dependent isocitrate ...
|
|
65411
|
Characterization of the physicochemical and catalytic properties of human heart NADP-dependent isocitrate ...
|
|
65412
|
Characterization of the physicochemical and catalytic properties of human heart NADP-dependent isocitrate ...
|
|
65413
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65414
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65415
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65416
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65417
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65418
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65419
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65420
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65421
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65422
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65423
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65424
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65425
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65426
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65427
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65428
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65429
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65430
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65431
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65432
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65433
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65434
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65435
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65436
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65437
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65438
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65439
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65440
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65441
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65442
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65443
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65444
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65445
|
Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and ...
|
|
65446
|
Evidence for the physiological role of a rhodanese-like protein for the biosynthesis of the molybdenum ...
|
|
65447
|
Roles of two conserved amino acid residues in the active site of galactose-1-phosphate uridylyltransferase: an ...
|
|
65448
|
Roles of two conserved amino acid residues in the active site of galactose-1-phosphate uridylyltransferase: an ...
|
|
65449
|
Roles of two conserved amino acid residues in the active site of galactose-1-phosphate uridylyltransferase: an ...
|
|
65450
|
Roles of two conserved amino acid residues in the active site of galactose-1-phosphate uridylyltransferase: an ...
|
|
65451
|
Roles of two conserved amino acid residues in the active site of galactose-1-phosphate uridylyltransferase: an ...
|
|
65452
|
Roles of two conserved amino acid residues in the active site of galactose-1-phosphate uridylyltransferase: an ...
|
|
65453
|
Roles of two conserved amino acid residues in the active site of galactose-1-phosphate uridylyltransferase: an ...
|
|
65454
|
Roles of two conserved amino acid residues in the active site of galactose-1-phosphate uridylyltransferase: an ...
|
|
65455
|
Roles of two conserved amino acid residues in the active site of galactose-1-phosphate uridylyltransferase: an ...
|
|
65456
|
An improved manual and semi-automatic assay for NADP-dependent isocitrate dehydrogenase activity, with a ...
|
|
65457
|
An improved manual and semi-automatic assay for NADP-dependent isocitrate dehydrogenase activity, with a ...
|
|
65458
|
An improved manual and semi-automatic assay for NADP-dependent isocitrate dehydrogenase activity, with a ...
|
|
65459
|
An improved manual and semi-automatic assay for NADP-dependent isocitrate dehydrogenase activity, with a ...
|
|
65460
|
An improved manual and semi-automatic assay for NADP-dependent isocitrate dehydrogenase activity, with a ...
|
|
65461
|
An improved manual and semi-automatic assay for NADP-dependent isocitrate dehydrogenase activity, with a ...
|
|
65462
|
An improved manual and semi-automatic assay for NADP-dependent isocitrate dehydrogenase activity, with a ...
|
|
65463
|
An improved manual and semi-automatic assay for NADP-dependent isocitrate dehydrogenase activity, with a ...
|
|
65464
|
An improved manual and semi-automatic assay for NADP-dependent isocitrate dehydrogenase activity, with a ...
|
|
65465
|
An α-1,6-and α-1,3-linked glucan produced by Leuconostoc citreum ABK-1 alternansucrase with nanoparticle and ...
|
|
65466
|
Characterization of two site-specifically mutated human dihydrolipoamide dehydrogenases (His-452----Gln and ...
|
|
65467
|
Characterization of two site-specifically mutated human dihydrolipoamide dehydrogenases (His-452----Gln and ...
|
|
65468
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65469
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65470
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65471
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65472
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65473
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65474
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65475
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65476
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65477
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65478
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65479
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65480
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65481
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65482
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65483
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65484
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65485
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65486
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65487
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65488
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65489
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65490
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65491
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65492
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65493
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65494
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65495
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65496
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65497
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65498
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65499
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65500
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65501
|
Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism
|
|
65502
|
Human heart TPN-specific isocitrate dehydrogenase. Purification by a rapid three-step procedure
|
|
65503
|
Human heart TPN-specific isocitrate dehydrogenase. Purification by a rapid three-step procedure
|
|
65504
|
Human heart TPN-specific isocitrate dehydrogenase. Purification by a rapid three-step procedure
|
|
65505
|
Human heart TPN-specific isocitrate dehydrogenase. Purification by a rapid three-step procedure
|
|
65506
|
Human heart TPN-specific isocitrate dehydrogenase. Purification by a rapid three-step procedure
|
|
65507
|
Each conserved active site tyr in the three subunits of human isocitrate dehydrogenase has a different ...
|
|
65508
|
Each conserved active site tyr in the three subunits of human isocitrate dehydrogenase has a different ...
|
|
65509
|
Each conserved active site tyr in the three subunits of human isocitrate dehydrogenase has a different ...
|
|
65510
|
Each conserved active site tyr in the three subunits of human isocitrate dehydrogenase has a different ...
|
|
65511
|
Each conserved active site tyr in the three subunits of human isocitrate dehydrogenase has a different ...
|
|
65512
|
Each conserved active site tyr in the three subunits of human isocitrate dehydrogenase has a different ...
|
|
65513
|
Each conserved active site tyr in the three subunits of human isocitrate dehydrogenase has a different ...
|
|
65514
|
Each conserved active site tyr in the three subunits of human isocitrate dehydrogenase has a different ...
|
|
65515
|
Each conserved active site tyr in the three subunits of human isocitrate dehydrogenase has a different ...
|
|
65516
|
Each conserved active site tyr in the three subunits of human isocitrate dehydrogenase has a different ...
|
|
65517
|
Each conserved active site tyr in the three subunits of human isocitrate dehydrogenase has a different ...
|
|
65518
|
Each conserved active site tyr in the three subunits of human isocitrate dehydrogenase has a different ...
|
|
65519
|
Each conserved active site tyr in the three subunits of human isocitrate dehydrogenase has a different ...
|
|
65520
|
Each conserved active site tyr in the three subunits of human isocitrate dehydrogenase has a different ...
|
|
65521
|
Each conserved active site tyr in the three subunits of human isocitrate dehydrogenase has a different ...
|
|
65522
|
Each conserved active site tyr in the three subunits of human isocitrate dehydrogenase has a different ...
|
|
65523
|
Each conserved active site tyr in the three subunits of human isocitrate dehydrogenase has a different ...
|
|
65524
|
Each conserved active site tyr in the three subunits of human isocitrate dehydrogenase has a different ...
|
|
65525
|
Each conserved active site tyr in the three subunits of human isocitrate dehydrogenase has a different ...
|
|
65526
|
Each conserved active site tyr in the three subunits of human isocitrate dehydrogenase has a different ...
|
|
65527
|
Each conserved active site tyr in the three subunits of human isocitrate dehydrogenase has a different ...
|
|
65528
|
Each conserved active site tyr in the three subunits of human isocitrate dehydrogenase has a different ...
|
|
65529
|
Each conserved active site tyr in the three subunits of human isocitrate dehydrogenase has a different ...
|
|
65530
|
Each conserved active site tyr in the three subunits of human isocitrate dehydrogenase has a different ...
|
|
65531
|
A novel nuclear human poly(A) polymerase (PAP), PAP gamma
|
|
65532
|
A novel nuclear human poly(A) polymerase (PAP), PAP gamma
|
|
65533
|
A novel nuclear human poly(A) polymerase (PAP), PAP gamma
|
|
65534
|
A novel nuclear human poly(A) polymerase (PAP), PAP gamma
|
|
65535
|
A novel nuclear human poly(A) polymerase (PAP), PAP gamma
|
|
65536
|
Archaeal RibL: a new FAD synthetase that is air sensitive
|
|
65537
|
Archaeal RibL: a new FAD synthetase that is air sensitive
|
|
65538
|
Archaeal RibL: a new FAD synthetase that is air sensitive
|
|
65539
|
The pentose catabolic pathway of the rice-blast fungus Magnaporthe oryzae involves a novel pentose reductase ...
|
|
65540
|
The pentose catabolic pathway of the rice-blast fungus Magnaporthe oryzae involves a novel pentose reductase ...
|
|
65541
|
Ribosylnicotinamide kinase domain of NadR protein: identification and implications in NAD biosynthesis
|
|
65542
|
Ribosylnicotinamide kinase domain of NadR protein: identification and implications in NAD biosynthesis
|
|
65543
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65544
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65545
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65546
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65547
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65548
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65549
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65550
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65551
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65552
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65553
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65554
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65555
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65556
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65557
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65558
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65559
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65560
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65561
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65562
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65563
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65564
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65565
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65566
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65567
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65568
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65569
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65570
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65571
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65572
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65573
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65574
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65575
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65576
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65577
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65578
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65579
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65580
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65581
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65582
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65583
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65584
|
Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor ...
|
|
65585
|
AtSUC3, a gene encoding a new Arabidopsis sucrose transporter, is expressed in cells adjacent to the vascular ...
|
|
65586
|
AtSUC3, a gene encoding a new Arabidopsis sucrose transporter, is expressed in cells adjacent to the vascular ...
|
|
65587
|
AtSUC3, a gene encoding a new Arabidopsis sucrose transporter, is expressed in cells adjacent to the vascular ...
|
|
65588
|
The Aspergillus niger D-xylulose kinase gene is co-expressed with genes encoding arabinan degrading enzymes, ...
|
|
65589
|
The Aspergillus niger D-xylulose kinase gene is co-expressed with genes encoding arabinan degrading enzymes, ...
|
|
65590
|
The Aspergillus niger D-xylulose kinase gene is co-expressed with genes encoding arabinan degrading enzymes, ...
|
|
65591
|
The Aspergillus niger D-xylulose kinase gene is co-expressed with genes encoding arabinan degrading enzymes, ...
|
|
65592
|
The Aspergillus niger D-xylulose kinase gene is co-expressed with genes encoding arabinan degrading enzymes, ...
|
|
65593
|
Structure and function of human xylulokinase, an enzyme with important roles in carbohydrate metabolism
|
|
65594
|
Structure and function of human xylulokinase, an enzyme with important roles in carbohydrate metabolism
|
|
65595
|
Purification and characterization of 6-phosphogluconate dehydrogenase from the wing-polymorphic cricket, ...
|
|
65596
|
Purification and characterization of 6-phosphogluconate dehydrogenase from the wing-polymorphic cricket, ...
|
|
65597
|
Purification and characterization of 6-phosphogluconate dehydrogenase from the wing-polymorphic cricket, ...
|
|
65598
|
Purification and characterization of 6-phosphogluconate dehydrogenase from the wing-polymorphic cricket, ...
|
|
65599
|
Purification and characterization of 6-phosphogluconate dehydrogenase from the wing-polymorphic cricket, ...
|
|
65600
|
Purification and characterization of 6-phosphogluconate dehydrogenase from the wing-polymorphic cricket, ...
|
|
65601
|
Purification and characterization of 6-phosphogluconate dehydrogenase from the wing-polymorphic cricket, ...
|
|
65602
|
Purification and characterization of 6-phosphogluconate dehydrogenase from the wing-polymorphic cricket, ...
|
|
65603
|
Purification and characterization of 6-phosphogluconate dehydrogenase from the wing-polymorphic cricket, ...
|
|
65604
|
Purification and characterization of 6-phosphogluconate dehydrogenase from the wing-polymorphic cricket, ...
|
|
65605
|
Purification and characterization of 6-phosphogluconate dehydrogenase from the wing-polymorphic cricket, ...
|
|
65606
|
Purification and characterization of 6-phosphogluconate dehydrogenase from the wing-polymorphic cricket, ...
|
|
65607
|
Purification and characterization of 6-phosphogluconate dehydrogenase from the wing-polymorphic cricket, ...
|
|
65608
|
Purification and characterization of 6-phosphogluconate dehydrogenase from the wing-polymorphic cricket, ...
|
|
65609
|
Purification and characterization of 6-phosphogluconate dehydrogenase from the wing-polymorphic cricket, ...
|
|
65610
|
Purification and characterization of 6-phosphogluconate dehydrogenase from the wing-polymorphic cricket, ...
|
|
65611
|
Expression, purification and characterization of recombinant phosphomannomutase and GDP-alpha-D-mannose ...
|
|
65612
|
Expression, purification and characterization of recombinant phosphomannomutase and GDP-alpha-D-mannose ...
|
|
65613
|
Expression, purification and characterization of recombinant phosphomannomutase and GDP-alpha-D-mannose ...
|
|
65614
|
Expression, purification and characterization of recombinant phosphomannomutase and GDP-alpha-D-mannose ...
|
|
65615
|
High-Throughput Screening of Coenzyme Preference Change of Thermophilic 6-Phosphogluconate Dehydrogenase from ...
|
|
65616
|
High-Throughput Screening of Coenzyme Preference Change of Thermophilic 6-Phosphogluconate Dehydrogenase from ...
|
|
65617
|
High-Throughput Screening of Coenzyme Preference Change of Thermophilic 6-Phosphogluconate Dehydrogenase from ...
|
|
65618
|
High-Throughput Screening of Coenzyme Preference Change of Thermophilic 6-Phosphogluconate Dehydrogenase from ...
|
|
65619
|
High-Throughput Screening of Coenzyme Preference Change of Thermophilic 6-Phosphogluconate Dehydrogenase from ...
|
|
65620
|
High-Throughput Screening of Coenzyme Preference Change of Thermophilic 6-Phosphogluconate Dehydrogenase from ...
|
|
65621
|
High-Throughput Screening of Coenzyme Preference Change of Thermophilic 6-Phosphogluconate Dehydrogenase from ...
|
|
65622
|
High-Throughput Screening of Coenzyme Preference Change of Thermophilic 6-Phosphogluconate Dehydrogenase from ...
|
|
65623
|
High-Throughput Screening of Coenzyme Preference Change of Thermophilic 6-Phosphogluconate Dehydrogenase from ...
|
|
65624
|
High-Throughput Screening of Coenzyme Preference Change of Thermophilic 6-Phosphogluconate Dehydrogenase from ...
|
|
65625
|
High-Throughput Screening of Coenzyme Preference Change of Thermophilic 6-Phosphogluconate Dehydrogenase from ...
|
|
65626
|
High-Throughput Screening of Coenzyme Preference Change of Thermophilic 6-Phosphogluconate Dehydrogenase from ...
|
|
65627
|
High-Throughput Screening of Coenzyme Preference Change of Thermophilic 6-Phosphogluconate Dehydrogenase from ...
|
|
65628
|
High-Throughput Screening of Coenzyme Preference Change of Thermophilic 6-Phosphogluconate Dehydrogenase from ...
|
|
65629
|
Coenzyme Engineering of a Hyperthermophilic 6-Phosphogluconate Dehydrogenase from NADP(+) to NAD(+) with Its ...
|
|
65630
|
Coenzyme Engineering of a Hyperthermophilic 6-Phosphogluconate Dehydrogenase from NADP(+) to NAD(+) with Its ...
|
|
65631
|
Coenzyme Engineering of a Hyperthermophilic 6-Phosphogluconate Dehydrogenase from NADP(+) to NAD(+) with Its ...
|
|
65632
|
Coenzyme Engineering of a Hyperthermophilic 6-Phosphogluconate Dehydrogenase from NADP(+) to NAD(+) with Its ...
|
|
65633
|
Coenzyme Engineering of a Hyperthermophilic 6-Phosphogluconate Dehydrogenase from NADP(+) to NAD(+) with Its ...
|
|
65634
|
Coenzyme Engineering of a Hyperthermophilic 6-Phosphogluconate Dehydrogenase from NADP(+) to NAD(+) with Its ...
|
|
65635
|
Coenzyme Engineering of a Hyperthermophilic 6-Phosphogluconate Dehydrogenase from NADP(+) to NAD(+) with Its ...
|
|
65636
|
Coenzyme Engineering of a Hyperthermophilic 6-Phosphogluconate Dehydrogenase from NADP(+) to NAD(+) with Its ...
|
|
65637
|
Coenzyme Engineering of a Hyperthermophilic 6-Phosphogluconate Dehydrogenase from NADP(+) to NAD(+) with Its ...
|
|
65638
|
6-Phosphogluconate dehydrogenase: the mechanism of action investigated by a comparison of the enzyme from ...
|
|
65639
|
6-Phosphogluconate dehydrogenase: the mechanism of action investigated by a comparison of the enzyme from ...
|
|
65640
|
6-Phosphogluconate dehydrogenase: the mechanism of action investigated by a comparison of the enzyme from ...
|
|
65641
|
6-Phosphogluconate dehydrogenase: the mechanism of action investigated by a comparison of the enzyme from ...
|
|
65642
|
6-Phosphogluconate dehydrogenase: the mechanism of action investigated by a comparison of the enzyme from ...
|
|
65643
|
6-Phosphogluconate dehydrogenase: the mechanism of action investigated by a comparison of the enzyme from ...
|
|
65644
|
6-Phosphogluconate dehydrogenase: the mechanism of action investigated by a comparison of the enzyme from ...
|
|
65645
|
6-Phosphogluconate dehydrogenase: the mechanism of action investigated by a comparison of the enzyme from ...
|
|
65646
|
6-Phosphogluconate dehydrogenase: the mechanism of action investigated by a comparison of the enzyme from ...
|
|
65647
|
6-Phosphogluconate dehydrogenase: the mechanism of action investigated by a comparison of the enzyme from ...
|
|
65648
|
Molecular cloning and functional characterization of three distinct N-methyltransferases involved in the ...
|
|
65649
|
Molecular cloning and functional characterization of three distinct N-methyltransferases involved in the ...
|
|
65650
|
Molecular cloning and functional characterization of three distinct N-methyltransferases involved in the ...
|
|
65651
|
Molecular cloning and functional characterization of three distinct N-methyltransferases involved in the ...
|
|
65652
|
Molecular cloning and functional characterization of three distinct N-methyltransferases involved in the ...
|
|
65653
|
Molecular cloning and functional characterization of three distinct N-methyltransferases involved in the ...
|
|
65654
|
Molecular cloning and functional characterization of three distinct N-methyltransferases involved in the ...
|
|
65655
|
Molecular cloning and functional characterization of three distinct N-methyltransferases involved in the ...
|
|
65656
|
Molecular cloning and functional characterization of three distinct N-methyltransferases involved in the ...
|
|
65657
|
Molecular cloning and functional characterization of three distinct N-methyltransferases involved in the ...
|
|
65658
|
Molecular cloning and functional characterization of three distinct N-methyltransferases involved in the ...
|
|
65659
|
Molecular cloning and functional characterization of three distinct N-methyltransferases involved in the ...
|
|
65660
|
Structural and functional analysis of a truncated form of Saccharomyces cerevisiae ATP sulfurylase: C-terminal ...
|
|
65661
|
Structural and functional analysis of a truncated form of Saccharomyces cerevisiae ATP sulfurylase: C-terminal ...
|
|
65662
|
Biochemical characterization of the GTP:adenosylcobinamide-phosphate guanylyltransferase (CobY) enzyme of the ...
|
|
65663
|
Biochemical characterization of the GTP:adenosylcobinamide-phosphate guanylyltransferase (CobY) enzyme of the ...
|
|
65664
|
Biochemical characterization of the GTP:adenosylcobinamide-phosphate guanylyltransferase (CobY) enzyme of the ...
|
|
65665
|
Biochemical characterization of the GTP:adenosylcobinamide-phosphate guanylyltransferase (CobY) enzyme of the ...
|
|
65666
|
The N-terminal domain of mammalian Lysyl-tRNA synthetase is a functional tRNA-binding domain
|
|
65667
|
The N-terminal domain of mammalian Lysyl-tRNA synthetase is a functional tRNA-binding domain
|
|
65668
|
The N-terminal domain of mammalian Lysyl-tRNA synthetase is a functional tRNA-binding domain
|
|
65669
|
The N-terminal domain of mammalian Lysyl-tRNA synthetase is a functional tRNA-binding domain
|
|
65670
|
The N-terminal domain of mammalian Lysyl-tRNA synthetase is a functional tRNA-binding domain
|
|
65671
|
The N-terminal domain of mammalian Lysyl-tRNA synthetase is a functional tRNA-binding domain
|
|
65672
|
The N-terminal domain of mammalian Lysyl-tRNA synthetase is a functional tRNA-binding domain
|
|
65673
|
The N-terminal domain of mammalian Lysyl-tRNA synthetase is a functional tRNA-binding domain
|
|
65674
|
The N-terminal domain of mammalian Lysyl-tRNA synthetase is a functional tRNA-binding domain
|
|
65675
|
The N-terminal domain of mammalian Lysyl-tRNA synthetase is a functional tRNA-binding domain
|
|
65676
|
The N-terminal domain of mammalian Lysyl-tRNA synthetase is a functional tRNA-binding domain
|
|
65677
|
The N-terminal domain of mammalian Lysyl-tRNA synthetase is a functional tRNA-binding domain
|
|
65678
|
The N-terminal domain of mammalian Lysyl-tRNA synthetase is a functional tRNA-binding domain
|
|
65679
|
The N-terminal domain of mammalian Lysyl-tRNA synthetase is a functional tRNA-binding domain
|
|
65680
|
The N-terminal domain of mammalian Lysyl-tRNA synthetase is a functional tRNA-binding domain
|
|
65681
|
The N-terminal domain of mammalian Lysyl-tRNA synthetase is a functional tRNA-binding domain
|
|
65682
|
The N-terminal domain of mammalian Lysyl-tRNA synthetase is a functional tRNA-binding domain
|
|
65683
|
The N-terminal domain of mammalian Lysyl-tRNA synthetase is a functional tRNA-binding domain
|
|
65684
|
The N-terminal domain of mammalian Lysyl-tRNA synthetase is a functional tRNA-binding domain
|
|
65685
|
The N-terminal domain of mammalian Lysyl-tRNA synthetase is a functional tRNA-binding domain
|
|
65686
|
The N-terminal domain of mammalian Lysyl-tRNA synthetase is a functional tRNA-binding domain
|
|
65687
|
Regulation of NAD synthesis by the trifunctional NadR protein of Salmonella enterica
|
|
65688
|
Regulation of NAD synthesis by the trifunctional NadR protein of Salmonella enterica
|
|
65689
|
Regulation of NAD synthesis by the trifunctional NadR protein of Salmonella enterica
|
|
65690
|
Regulation of NAD synthesis by the trifunctional NadR protein of Salmonella enterica
|
|
65691
|
Regulation of NAD synthesis by the trifunctional NadR protein of Salmonella enterica
|
|
65692
|
Regulation of NAD synthesis by the trifunctional NadR protein of Salmonella enterica
|
|
65693
|
Targeted metabolite and transcript profiling for elucidating enzyme function: isolation of novel ...
|
|
65694
|
Targeted metabolite and transcript profiling for elucidating enzyme function: isolation of novel ...
|
|
65695
|
Targeted metabolite and transcript profiling for elucidating enzyme function: isolation of novel ...
|
|
65696
|
Targeted metabolite and transcript profiling for elucidating enzyme function: isolation of novel ...
|
|
65697
|
Targeted metabolite and transcript profiling for elucidating enzyme function: isolation of novel ...
|
|
65698
|
Targeted metabolite and transcript profiling for elucidating enzyme function: isolation of novel ...
|
|
65699
|
Second messenger signalling governs Escherichia coli biofilm induction upon ribosomal stress
|
|
65700
|
Second messenger signalling governs Escherichia coli biofilm induction upon ribosomal stress
|
|
65701
|
Isolation and characterization of a digoxin transporter and its rat homologue expressed in the kidney.
|
|
65702
|
Isolation and characterization of a digoxin transporter and its rat homologue expressed in the kidney.
|
|
65703
|
Isolation and characterization of a digoxin transporter and its rat homologue expressed in the kidney.
|
|
65704
|
Isolation and characterization of a digoxin transporter and its rat homologue expressed in the kidney.
|
|
65705
|
Isolation and characterization of a digoxin transporter and its rat homologue expressed in the kidney.
|
|
65706
|
Isolation and characterization of a digoxin transporter and its rat homologue expressed in the kidney.
|
|
65707
|
Isolation and characterization of a digoxin transporter and its rat homologue expressed in the kidney.
|
|
65708
|
Isolation and characterization of a digoxin transporter and its rat homologue expressed in the kidney.
|
|
65709
|
Isolation and characterization of a digoxin transporter and its rat homologue expressed in the kidney.
|
|
65710
|
Isolation and characterization of a digoxin transporter and its rat homologue expressed in the kidney.
|
|
65711
|
Isolation and characterization of a digoxin transporter and its rat homologue expressed in the kidney.
|
|
65712
|
Isolation and characterization of a digoxin transporter and its rat homologue expressed in the kidney.
|
|
65713
|
Inhibition of the Na+/dicarboxylate cotransporter by anthranilic acid derivatives
|
|
65714
|
Inhibition of the Na+/dicarboxylate cotransporter by anthranilic acid derivatives
|
|
65715
|
Inhibition of the Na+/dicarboxylate cotransporter by anthranilic acid derivatives
|
|
65716
|
Identification and characterization of a strict and a promiscuous N-acetylglucosamine-1-P uridylyltransferase ...
|
|
65717
|
Identification and characterization of a strict and a promiscuous N-acetylglucosamine-1-P uridylyltransferase ...
|
|
65718
|
Identification and characterization of a strict and a promiscuous N-acetylglucosamine-1-P uridylyltransferase ...
|
|
65719
|
Identification and characterization of a strict and a promiscuous N-acetylglucosamine-1-P uridylyltransferase ...
|
|
65720
|
Identification and characterization of a strict and a promiscuous N-acetylglucosamine-1-P uridylyltransferase ...
|
|
65721
|
Identification and characterization of a strict and a promiscuous N-acetylglucosamine-1-P uridylyltransferase ...
|
|
65722
|
Identification and characterization of a strict and a promiscuous N-acetylglucosamine-1-P uridylyltransferase ...
|
|
65723
|
Identification and characterization of a strict and a promiscuous N-acetylglucosamine-1-P uridylyltransferase ...
|
|
65724
|
Identification and characterization of a strict and a promiscuous N-acetylglucosamine-1-P uridylyltransferase ...
|
|
65725
|
Expression cloning and characterization of human 17 beta-hydroxysteroid dehydrogenase type 2, a microsomal ...
|
|
65726
|
Expression cloning and characterization of human 17 beta-hydroxysteroid dehydrogenase type 2, a microsomal ...
|
|
65727
|
Expression cloning and characterization of human 17 beta-hydroxysteroid dehydrogenase type 2, a microsomal ...
|
|
65728
|
Expression cloning and characterization of human 17 beta-hydroxysteroid dehydrogenase type 2, a microsomal ...
|
|
65729
|
Expression cloning and characterization of human 17 beta-hydroxysteroid dehydrogenase type 2, a microsomal ...
|
|
65730
|
Expression cloning and characterization of human 17 beta-hydroxysteroid dehydrogenase type 2, a microsomal ...
|
|
65731
|
Characterization and in vitro expression of arginine kinase gene in the invasive western flower thrips, ...
|
|
65732
|
Gamma-Glutamyltransferase: kinetic properties and assay conditions when gamma-glutamyl-4-nitroanilide and its ...
|
|
65733
|
Gamma-Glutamyltransferase: kinetic properties and assay conditions when gamma-glutamyl-4-nitroanilide and its ...
|
|
65734
|
Cloning and characterization of a pyrethroid pesticide decomposing esterase gene, Est3385, from ...
|
|
65735
|
Investigation of the proton relay system operative in human cystosolic aminopeptidase P.
|
|
65736
|
Investigation of the proton relay system operative in human cystosolic aminopeptidase P.
|
|
65737
|
Investigation of the proton relay system operative in human cystosolic aminopeptidase P.
|
|
65738
|
Investigation of the proton relay system operative in human cystosolic aminopeptidase P.
|
|
65739
|
Investigation of the proton relay system operative in human cystosolic aminopeptidase P.
|
|
65740
|
Investigation of the proton relay system operative in human cystosolic aminopeptidase P.
|
|
65741
|
Investigation of the proton relay system operative in human cystosolic aminopeptidase P.
|
|
65742
|
Investigation of the proton relay system operative in human cystosolic aminopeptidase P.
|
|
65743
|
Investigation of the proton relay system operative in human cystosolic aminopeptidase P.
|
|
65744
|
Malate dehydrogenase. Kinetic studies with meso-tartrate and 2-keto-3-hydroxysuccinate, comparison of the ...
|
|
65745
|
Malate dehydrogenase. Kinetic studies with meso-tartrate and 2-keto-3-hydroxysuccinate, comparison of the ...
|
|
65746
|
Malate dehydrogenase. Kinetic studies with meso-tartrate and 2-keto-3-hydroxysuccinate, comparison of the ...
|
|
65747
|
Malate dehydrogenase. Kinetic studies with meso-tartrate and 2-keto-3-hydroxysuccinate, comparison of the ...
|
|
65748
|
Malate dehydrogenase. Kinetic studies with meso-tartrate and 2-keto-3-hydroxysuccinate, comparison of the ...
|
|
65749
|
Malate dehydrogenase. Kinetic studies with meso-tartrate and 2-keto-3-hydroxysuccinate, comparison of the ...
|
|
65750
|
Malate dehydrogenase. Kinetic studies with meso-tartrate and 2-keto-3-hydroxysuccinate, comparison of the ...
|
|
65751
|
Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis.
|
|
65752
|
Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis.
|
|
65753
|
Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis.
|
|
65754
|
Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis.
|
|
65755
|
Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis.
|
|
65756
|
Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis.
|
|
65757
|
Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis.
|
|
65758
|
Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis.
|
|
65759
|
Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis.
|
|
65760
|
Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis.
|
|
65761
|
Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis.
|
|
65762
|
Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis.
|
|
65763
|
Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis.
|
|
65764
|
Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis.
|
|
65765
|
Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis.
|
|
65766
|
Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis.
|
|
65767
|
Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis.
|
|
65768
|
Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis.
|
|
65769
|
Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis.
|
|
65770
|
Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis.
|
|
65771
|
Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis.
|
|
65772
|
Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis.
|
|
65773
|
Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis.
|
|
65774
|
Identification of enzymes acting on alpha-glycated amino acids in Bacillus subtilis.
|
|
65775
|
Ferredoxin-NADP+ reductase from Plasmodium falciparum undergoes NADP+-dependent dimerization and inactivation: ...
|
|
65776
|
Ferredoxin-NADP+ reductase from Plasmodium falciparum undergoes NADP+-dependent dimerization and inactivation: ...
|
|
65777
|
Ferredoxin-NADP+ reductase from Plasmodium falciparum undergoes NADP+-dependent dimerization and inactivation: ...
|
|
65778
|
Ferredoxin-NADP+ reductase from Plasmodium falciparum undergoes NADP+-dependent dimerization and inactivation: ...
|
|
65779
|
Ferredoxin-NADP+ reductase from Plasmodium falciparum undergoes NADP+-dependent dimerization and inactivation: ...
|
|
65780
|
Ferredoxin-NADP+ reductase from Plasmodium falciparum undergoes NADP+-dependent dimerization and inactivation: ...
|
|
65781
|
Ferredoxin-NADP+ reductase from Plasmodium falciparum undergoes NADP+-dependent dimerization and inactivation: ...
|
|
65782
|
The heterodimeric primase from the euryarchaeon Pyrococcus abyssi: a multifunctional enzyme for initiation and ...
|
|
65783
|
The heterodimeric primase from the euryarchaeon Pyrococcus abyssi: a multifunctional enzyme for initiation and ...
|
|
65784
|
The heterodimeric primase from the euryarchaeon Pyrococcus abyssi: a multifunctional enzyme for initiation and ...
|
|
65785
|
Promiscuous DNA synthesis by human DNA polymerase ?
|
|
65786
|
Promiscuous DNA synthesis by human DNA polymerase ?
|
|
65787
|
Promiscuous DNA synthesis by human DNA polymerase ?
|
|
65788
|
Synthesis and characterization of cross linked enzyme aggregates of serine hydroxyl methyltransferase from ...
|
|
65789
|
Synthesis and characterization of cross linked enzyme aggregates of serine hydroxyl methyltransferase from ...
|
|
65790
|
Crystal structure and functional analysis identify the P-loop containing protein YFH7 of Saccharomyces ...
|
|
65791
|
Crystal structure and functional analysis identify the P-loop containing protein YFH7 of Saccharomyces ...
|
|
65792
|
PknB-mediated phosphorylation of a novel substrate, N-acetylglucosamine-1-phosphate uridyltransferase, ...
|
|
65793
|
PknB-mediated phosphorylation of a novel substrate, N-acetylglucosamine-1-phosphate uridyltransferase, ...
|
|
65794
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65795
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65796
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65797
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65798
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65799
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65800
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65801
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65802
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65803
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65804
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65805
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65806
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65807
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65808
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65809
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65810
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65811
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65812
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65813
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65814
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65815
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65816
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65817
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65818
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65819
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65820
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65821
|
Characterization of the functional role of Asp141, Asp194, and Asp464 residues in the Mn2+-L-malate binding of ...
|
|
65822
|
The E2 domain of OdhA of Corynebacterium glutamicum has succinyltransferase activity dependent on lipoyl ...
|
|
65823
|
The E2 domain of OdhA of Corynebacterium glutamicum has succinyltransferase activity dependent on lipoyl ...
|
|
65824
|
The E2 domain of OdhA of Corynebacterium glutamicum has succinyltransferase activity dependent on lipoyl ...
|
|
65825
|
The E2 domain of OdhA of Corynebacterium glutamicum has succinyltransferase activity dependent on lipoyl ...
|
|
65826
|
The E2 domain of OdhA of Corynebacterium glutamicum has succinyltransferase activity dependent on lipoyl ...
|
|
65827
|
The E2 domain of OdhA of Corynebacterium glutamicum has succinyltransferase activity dependent on lipoyl ...
|
|
65828
|
The E2 domain of OdhA of Corynebacterium glutamicum has succinyltransferase activity dependent on lipoyl ...
|
|
65829
|
The E2 domain of OdhA of Corynebacterium glutamicum has succinyltransferase activity dependent on lipoyl ...
|
|
65830
|
Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites
|
|
65831
|
Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites
|
|
65832
|
Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites
|
|
65833
|
Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites
|
|
65834
|
Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites
|
|
65835
|
Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites
|
|
65836
|
Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites
|
|
65837
|
Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites
|
|
65838
|
Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites
|
|
65839
|
Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites
|
|
65840
|
Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites
|
|
65841
|
Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites
|
|
65842
|
Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites
|
|
65843
|
Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites
|
|
65844
|
Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites
|
|
65845
|
Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites
|
|
65846
|
Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites
|
|
65847
|
Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites
|
|
65848
|
Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites
|
|
65849
|
Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites
|
|
65850
|
Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites
|
|
65851
|
Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites
|
|
65852
|
Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites
|
|
65853
|
Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites
|
|
65854
|
Structural basis for dimerization and activity of human PAPD1, a noncanonical poly(A) polymerase
|
|
65855
|
Structural basis for dimerization and activity of human PAPD1, a noncanonical poly(A) polymerase
|
|
65856
|
Non-active site mutation (Q123A) in New Delhi metallo-beta-lactamase (NDM-1) enhanced its enzyme activity.
|
|
65857
|
Non-active site mutation (Q123A) in New Delhi metallo-beta-lactamase (NDM-1) enhanced its enzyme activity.
|
|
65858
|
Non-active site mutation (Q123A) in New Delhi metallo-beta-lactamase (NDM-1) enhanced its enzyme activity.
|
|
65859
|
Non-active site mutation (Q123A) in New Delhi metallo-beta-lactamase (NDM-1) enhanced its enzyme activity.
|
|
65860
|
Non-active site mutation (Q123A) in New Delhi metallo-beta-lactamase (NDM-1) enhanced its enzyme activity.
|
|
65861
|
Non-active site mutation (Q123A) in New Delhi metallo-beta-lactamase (NDM-1) enhanced its enzyme activity.
|
|
65862
|
Non-active site mutation (Q123A) in New Delhi metallo-beta-lactamase (NDM-1) enhanced its enzyme activity.
|
|
65863
|
Non-active site mutation (Q123A) in New Delhi metallo-beta-lactamase (NDM-1) enhanced its enzyme activity.
|
|
65864
|
Non-active site mutation (Q123A) in New Delhi metallo-beta-lactamase (NDM-1) enhanced its enzyme activity.
|
|
65865
|
Non-active site mutation (Q123A) in New Delhi metallo-beta-lactamase (NDM-1) enhanced its enzyme activity.
|
|
65866
|
Non-active site mutation (Q123A) in New Delhi metallo-beta-lactamase (NDM-1) enhanced its enzyme activity.
|
|
65867
|
Non-active site mutation (Q123A) in New Delhi metallo-beta-lactamase (NDM-1) enhanced its enzyme activity.
|
|
65868
|
Non-active site mutation (Q123A) in New Delhi metallo-beta-lactamase (NDM-1) enhanced its enzyme activity.
|
|
65869
|
Non-active site mutation (Q123A) in New Delhi metallo-beta-lactamase (NDM-1) enhanced its enzyme activity.
|
|
65870
|
New Insights into SN-38 Glucuronidation: Evidence for the Important Role of UDP Glucuronosyltransferase 1A9.
|
|
65871
|
New Insights into SN-38 Glucuronidation: Evidence for the Important Role of UDP Glucuronosyltransferase 1A9.
|
|
65872
|
New Insights into SN-38 Glucuronidation: Evidence for the Important Role of UDP Glucuronosyltransferase 1A9.
|
|
65873
|
New Insights into SN-38 Glucuronidation: Evidence for the Important Role of UDP Glucuronosyltransferase 1A9.
|
|
65874
|
New Insights into SN-38 Glucuronidation: Evidence for the Important Role of UDP Glucuronosyltransferase 1A9.
|
|
65875
|
New Insights into SN-38 Glucuronidation: Evidence for the Important Role of UDP Glucuronosyltransferase 1A9.
|
|
65876
|
Development of a selective and highly sensitive fluorescence assay for nucleoside triphosphate ...
|
|
65877
|
Development of a selective and highly sensitive fluorescence assay for nucleoside triphosphate ...
|
|
65878
|
Development of a selective and highly sensitive fluorescence assay for nucleoside triphosphate ...
|
|
65879
|
Development of a selective and highly sensitive fluorescence assay for nucleoside triphosphate ...
|
|
65880
|
Development of a selective and highly sensitive fluorescence assay for nucleoside triphosphate ...
|
|
65881
|
Development of a selective and highly sensitive fluorescence assay for nucleoside triphosphate ...
|
|
65882
|
Development of a selective and highly sensitive fluorescence assay for nucleoside triphosphate ...
|
|
65883
|
Development of a selective and highly sensitive fluorescence assay for nucleoside triphosphate ...
|
|
65884
|
Development of a selective and highly sensitive fluorescence assay for nucleoside triphosphate ...
|
|
65885
|
Development of a selective and highly sensitive fluorescence assay for nucleoside triphosphate ...
|
|
65886
|
Development of a selective and highly sensitive fluorescence assay for nucleoside triphosphate ...
|
|
65887
|
Development of a selective and highly sensitive fluorescence assay for nucleoside triphosphate ...
|
|
65888
|
Development of a selective and highly sensitive fluorescence assay for nucleoside triphosphate ...
|
|
65889
|
Development of a selective and highly sensitive fluorescence assay for nucleoside triphosphate ...
|
|
65890
|
Development of a selective and highly sensitive fluorescence assay for nucleoside triphosphate ...
|
|
65891
|
Development of a selective and highly sensitive fluorescence assay for nucleoside triphosphate ...
|
|
65892
|
Development of a selective and highly sensitive fluorescence assay for nucleoside triphosphate ...
|
|
65893
|
Development of a selective and highly sensitive fluorescence assay for nucleoside triphosphate ...
|
|
65894
|
Development of a selective and highly sensitive fluorescence assay for nucleoside triphosphate ...
|
|
65895
|
Development of a selective and highly sensitive fluorescence assay for nucleoside triphosphate ...
|
|
65896
|
KINETIC STUDIES OF GLUTAMIC OXALOACETIC TRANSAMINASE ISOZYMES.
|
|
65897
|
KINETIC STUDIES OF GLUTAMIC OXALOACETIC TRANSAMINASE ISOZYMES.
|
|
65898
|
MocA is a specific cytidylyltransferase involved in molybdopterin cytosine dinucleotide biosynthesis in ...
|
|
65899
|
MocA is a specific cytidylyltransferase involved in molybdopterin cytosine dinucleotide biosynthesis in ...
|
|
65900
|
MocA is a specific cytidylyltransferase involved in molybdopterin cytosine dinucleotide biosynthesis in ...
|
|
65901
|
MocA is a specific cytidylyltransferase involved in molybdopterin cytosine dinucleotide biosynthesis in ...
|
|
65902
|
MocA is a specific cytidylyltransferase involved in molybdopterin cytosine dinucleotide biosynthesis in ...
|
|
65903
|
MocA is a specific cytidylyltransferase involved in molybdopterin cytosine dinucleotide biosynthesis in ...
|
|
65904
|
MocA is a specific cytidylyltransferase involved in molybdopterin cytosine dinucleotide biosynthesis in ...
|
|
65905
|
MocA is a specific cytidylyltransferase involved in molybdopterin cytosine dinucleotide biosynthesis in ...
|
|
65906
|
MocA is a specific cytidylyltransferase involved in molybdopterin cytosine dinucleotide biosynthesis in ...
|
|
65907
|
MocA is a specific cytidylyltransferase involved in molybdopterin cytosine dinucleotide biosynthesis in ...
|
|
65908
|
MocA is a specific cytidylyltransferase involved in molybdopterin cytosine dinucleotide biosynthesis in ...
|
|
65909
|
MocA is a specific cytidylyltransferase involved in molybdopterin cytosine dinucleotide biosynthesis in ...
|
|
65910
|
MocA is a specific cytidylyltransferase involved in molybdopterin cytosine dinucleotide biosynthesis in ...
|
|
65911
|
MocA is a specific cytidylyltransferase involved in molybdopterin cytosine dinucleotide biosynthesis in ...
|
|
65912
|
Interaction of alpha-lipoic acid enantiomers and homologues with the enzyme components of the mammalian ...
|
|
65913
|
Interaction of alpha-lipoic acid enantiomers and homologues with the enzyme components of the mammalian ...
|
|
65914
|
Interaction of alpha-lipoic acid enantiomers and homologues with the enzyme components of the mammalian ...
|
|
65915
|
Interaction of alpha-lipoic acid enantiomers and homologues with the enzyme components of the mammalian ...
|
|
65916
|
Interaction of alpha-lipoic acid enantiomers and homologues with the enzyme components of the mammalian ...
|
|
65917
|
Interaction of alpha-lipoic acid enantiomers and homologues with the enzyme components of the mammalian ...
|
|
65918
|
Interaction of alpha-lipoic acid enantiomers and homologues with the enzyme components of the mammalian ...
|
|
65919
|
Interaction of alpha-lipoic acid enantiomers and homologues with the enzyme components of the mammalian ...
|
|
65920
|
Interaction of alpha-lipoic acid enantiomers and homologues with the enzyme components of the mammalian ...
|
|
65921
|
Interaction of alpha-lipoic acid enantiomers and homologues with the enzyme components of the mammalian ...
|
|
65922
|
Interaction of alpha-lipoic acid enantiomers and homologues with the enzyme components of the mammalian ...
|
|
65923
|
Interaction of alpha-lipoic acid enantiomers and homologues with the enzyme components of the mammalian ...
|
|
65924
|
Heterologous expression of Aspergillus niger beta-D-xylosidase (XlnD): characterization on lignocellulosic ...
|
|
65925
|
Heterologous expression of Aspergillus niger beta-D-xylosidase (XlnD): characterization on lignocellulosic ...
|
|
65926
|
Degradation of xylan to D-xylose by recombinant Saccharomyces cerevisiae coexpressing the Aspergillus niger ...
|
|
65927
|
Degradation of xylan to D-xylose by recombinant Saccharomyces cerevisiae coexpressing the Aspergillus niger ...
|
|
65928
|
Galactose-1-phosphate uridylyltransferase: rate studies confirming a uridylyl-enzyme intermediate on the ...
|
|
65929
|
Galactose-1-phosphate uridylyltransferase: rate studies confirming a uridylyl-enzyme intermediate on the ...
|
|
65930
|
Galactose-1-phosphate uridylyltransferase: rate studies confirming a uridylyl-enzyme intermediate on the ...
|
|
65931
|
Galactose-1-phosphate uridylyltransferase: rate studies confirming a uridylyl-enzyme intermediate on the ...
|
|
65932
|
Galactose-1-phosphate uridylyltransferase: rate studies confirming a uridylyl-enzyme intermediate on the ...
|
|
65933
|
Galactose-1-phosphate uridylyltransferase: rate studies confirming a uridylyl-enzyme intermediate on the ...
|
|
65934
|
Galactose-1-phosphate uridylyltransferase: rate studies confirming a uridylyl-enzyme intermediate on the ...
|
|
65935
|
Galactose-1-phosphate uridylyltransferase: rate studies confirming a uridylyl-enzyme intermediate on the ...
|
|
65936
|
Galactose-1-phosphate uridylyltransferase: rate studies confirming a uridylyl-enzyme intermediate on the ...
|
|
65937
|
Galactose-1-phosphate uridylyltransferase: rate studies confirming a uridylyl-enzyme intermediate on the ...
|
|
65938
|
Characteristics of thermostable endoxylanase and ?-xylosidase of the extremely thermophilic bacterium ...
|
|
65939
|
Characteristics of thermostable endoxylanase and ?-xylosidase of the extremely thermophilic bacterium ...
|
|
65940
|
Talaromyces thermophilus beta-D-xylosidase: purification, characterization and xylobiose synthesis
|
|
65941
|
Mode of action and properties of the beta-xylosidases from Talaromyces emersonii and Trichoderma reesei
|
|
65942
|
Mode of action and properties of the beta-xylosidases from Talaromyces emersonii and Trichoderma reesei
|
|
65943
|
Mode of action and properties of the beta-xylosidases from Talaromyces emersonii and Trichoderma reesei
|
|
65944
|
Mode of action and properties of the beta-xylosidases from Talaromyces emersonii and Trichoderma reesei
|
|
65945
|
Mode of action and properties of the beta-xylosidases from Talaromyces emersonii and Trichoderma reesei
|
|
65946
|
Mode of action and properties of the beta-xylosidases from Talaromyces emersonii and Trichoderma reesei
|
|
65947
|
Mode of action and properties of the beta-xylosidases from Talaromyces emersonii and Trichoderma reesei
|
|
65948
|
Mode of action and properties of the beta-xylosidases from Talaromyces emersonii and Trichoderma reesei
|
|
65949
|
Mode of action and properties of the beta-xylosidases from Talaromyces emersonii and Trichoderma reesei
|
|
65950
|
Mode of action and properties of the beta-xylosidases from Talaromyces emersonii and Trichoderma reesei
|
|
65951
|
Mode of action and properties of the beta-xylosidases from Talaromyces emersonii and Trichoderma reesei
|
|
65952
|
Mode of action and properties of the beta-xylosidases from Talaromyces emersonii and Trichoderma reesei
|
|
65953
|
Mode of action and properties of the beta-xylosidases from Talaromyces emersonii and Trichoderma reesei
|
|
65954
|
Mode of action and properties of the beta-xylosidases from Talaromyces emersonii and Trichoderma reesei
|
|
65955
|
Fumarate reductase: a target for therapeutic intervention against Helicobacter pylori.
|
|
65956
|
Fumarate reductase: a target for therapeutic intervention against Helicobacter pylori.
|
|
65957
|
Fumarate reductase: a target for therapeutic intervention against Helicobacter pylori.
|
|
65958
|
Fumarate reductase: a target for therapeutic intervention against Helicobacter pylori.
|
|
65959
|
Determination of the mechanism and kinetic constants for hog kidney gamma-glutamyltransferase.
|
|
65960
|
Determination of the mechanism and kinetic constants for hog kidney gamma-glutamyltransferase.
|
|
65961
|
Uncovering divergent evolution of α/β-hydrolases: a surprising residue substitution needed to convert Hevea ...
|
|
65962
|
Uncovering divergent evolution of α/β-hydrolases: a surprising residue substitution needed to convert Hevea ...
|
|
65963
|
Uncovering divergent evolution of α/β-hydrolases: a surprising residue substitution needed to convert Hevea ...
|
|
65964
|
Uncovering divergent evolution of α/β-hydrolases: a surprising residue substitution needed to convert Hevea ...
|
|
65965
|
Uncovering divergent evolution of α/β-hydrolases: a surprising residue substitution needed to convert Hevea ...
|
|
65966
|
Uncovering divergent evolution of α/β-hydrolases: a surprising residue substitution needed to convert Hevea ...
|
|
65967
|
Uncovering divergent evolution of α/β-hydrolases: a surprising residue substitution needed to convert Hevea ...
|
|
65968
|
Uncovering divergent evolution of α/β-hydrolases: a surprising residue substitution needed to convert Hevea ...
|
|
65969
|
Uncovering divergent evolution of α/β-hydrolases: a surprising residue substitution needed to convert Hevea ...
|
|
65970
|
Uncovering divergent evolution of α/β-hydrolases: a surprising residue substitution needed to convert Hevea ...
|
|
65971
|
Uncovering divergent evolution of α/β-hydrolases: a surprising residue substitution needed to convert Hevea ...
|
|
65972
|
Uncovering divergent evolution of α/β-hydrolases: a surprising residue substitution needed to convert Hevea ...
|
|
65973
|
Uncovering divergent evolution of α/β-hydrolases: a surprising residue substitution needed to convert Hevea ...
|
|
65974
|
Uncovering divergent evolution of α/β-hydrolases: a surprising residue substitution needed to convert Hevea ...
|
|
65975
|
Uncovering divergent evolution of α/β-hydrolases: a surprising residue substitution needed to convert Hevea ...
|
|
65976
|
Uncovering divergent evolution of α/β-hydrolases: a surprising residue substitution needed to convert Hevea ...
|
|
65977
|
Uncovering divergent evolution of α/β-hydrolases: a surprising residue substitution needed to convert Hevea ...
|
|
65978
|
Uncovering divergent evolution of α/β-hydrolases: a surprising residue substitution needed to convert Hevea ...
|
|
65979
|
Uncovering divergent evolution of α/β-hydrolases: a surprising residue substitution needed to convert Hevea ...
|
|
65980
|
Molybdopterin dinucleotide biosynthesis in Escherichia coli: identification of amino acid residues of ...
|
|
65981
|
Molybdopterin dinucleotide biosynthesis in Escherichia coli: identification of amino acid residues of ...
|
|
65982
|
Molybdopterin dinucleotide biosynthesis in Escherichia coli: identification of amino acid residues of ...
|
|
65983
|
Molybdopterin dinucleotide biosynthesis in Escherichia coli: identification of amino acid residues of ...
|
|
65984
|
Molybdopterin dinucleotide biosynthesis in Escherichia coli: identification of amino acid residues of ...
|
|
65985
|
Molybdopterin dinucleotide biosynthesis in Escherichia coli: identification of amino acid residues of ...
|
|
65986
|
Molybdopterin dinucleotide biosynthesis in Escherichia coli: identification of amino acid residues of ...
|
|
65987
|
Molybdopterin dinucleotide biosynthesis in Escherichia coli: identification of amino acid residues of ...
|
|
65988
|
Molybdopterin dinucleotide biosynthesis in Escherichia coli: identification of amino acid residues of ...
|
|
65989
|
Molybdopterin dinucleotide biosynthesis in Escherichia coli: identification of amino acid residues of ...
|
|
65990
|
Molybdopterin dinucleotide biosynthesis in Escherichia coli: identification of amino acid residues of ...
|
|
65991
|
Molybdopterin dinucleotide biosynthesis in Escherichia coli: identification of amino acid residues of ...
|
|
65992
|
Molybdopterin dinucleotide biosynthesis in Escherichia coli: identification of amino acid residues of ...
|
|
65993
|
Molybdopterin dinucleotide biosynthesis in Escherichia coli: identification of amino acid residues of ...
|
|
65994
|
Molybdopterin dinucleotide biosynthesis in Escherichia coli: identification of amino acid residues of ...
|
|
65995
|
Molybdopterin dinucleotide biosynthesis in Escherichia coli: identification of amino acid residues of ...
|
|
65996
|
Molybdopterin dinucleotide biosynthesis in Escherichia coli: identification of amino acid residues of ...
|
|
65997
|
Molybdopterin dinucleotide biosynthesis in Escherichia coli: identification of amino acid residues of ...
|
|
65998
|
Molybdopterin dinucleotide biosynthesis in Escherichia coli: identification of amino acid residues of ...
|
|
65999
|
Molybdopterin dinucleotide biosynthesis in Escherichia coli: identification of amino acid residues of ...
|
|
66000
|
Molybdopterin dinucleotide biosynthesis in Escherichia coli: identification of amino acid residues of ...
|





 If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.
If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.