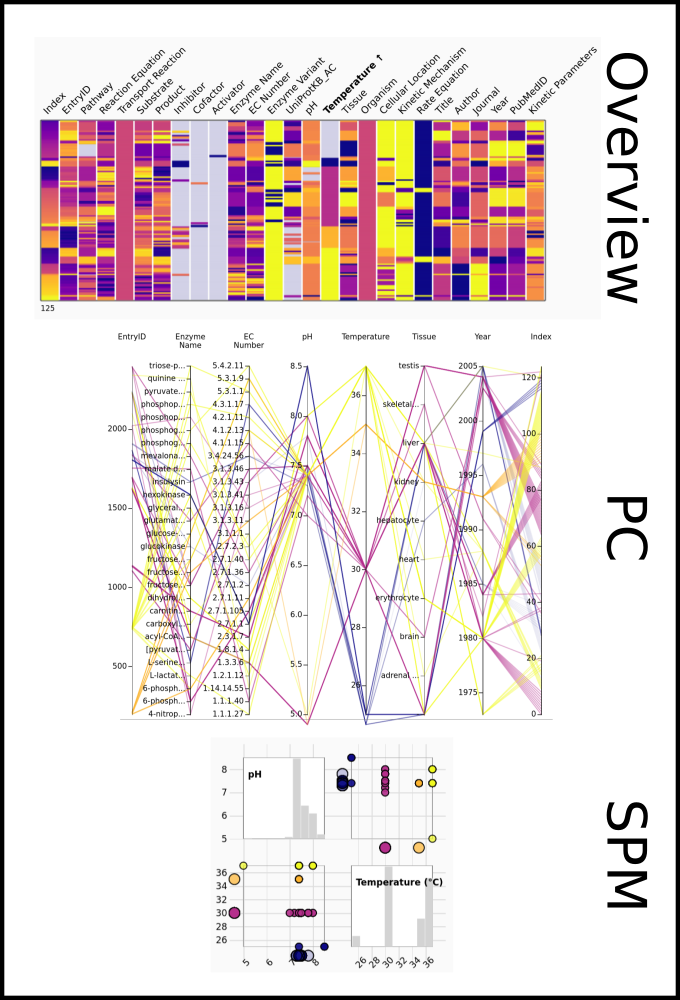
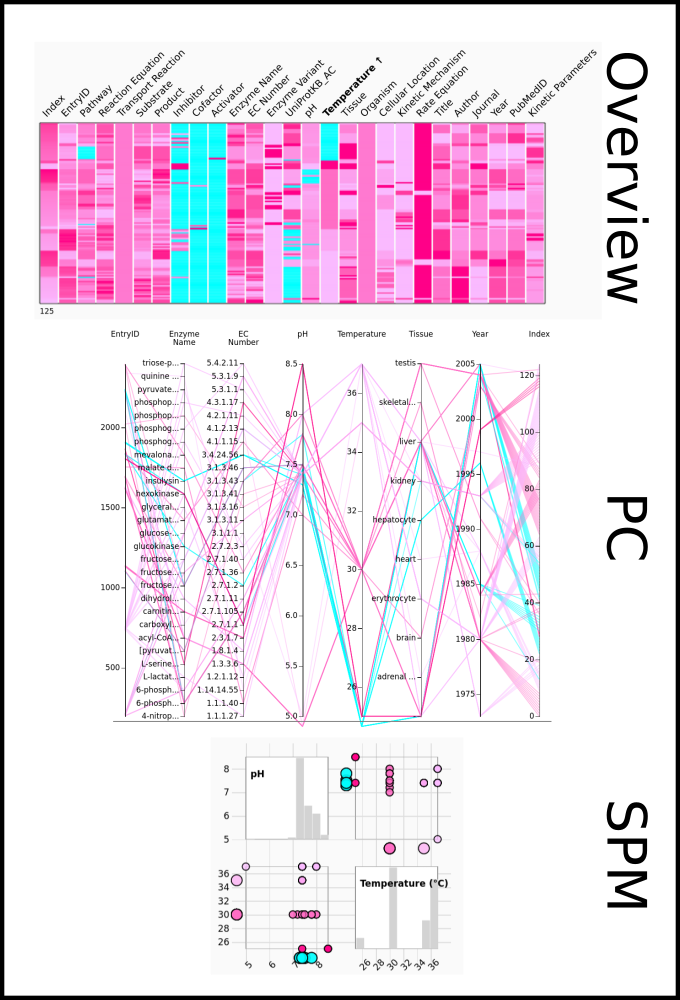
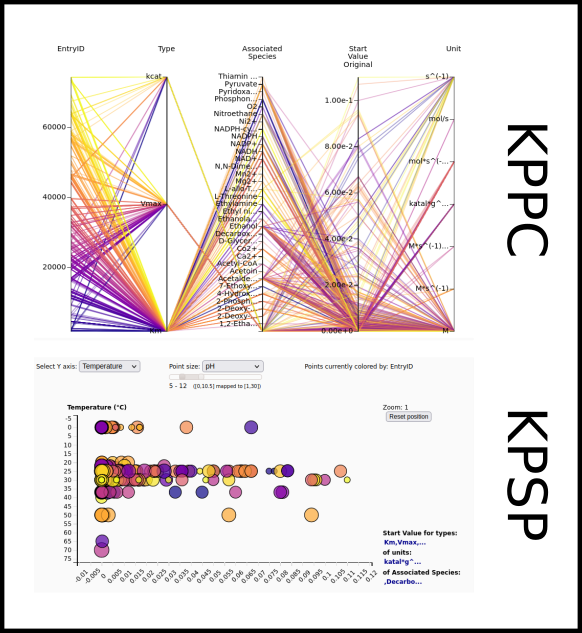
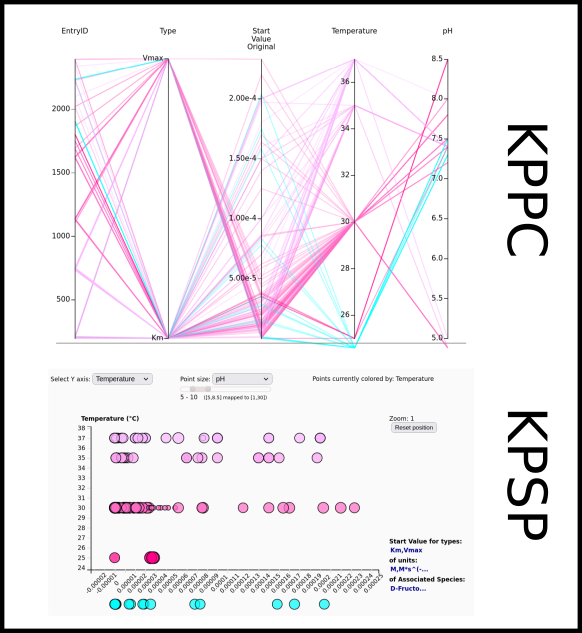
To improve SABIO-RK data interoperability, semantic markup was added to web pages as described and defined by
This structured information makes it easier to discover, collate and analyse our data.
|
53001
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
53002
|
Purification and characterization of the F1 ATPase from Bacillus subtilis and its uncoupler-resistant mutant ...
|
|
53003
|
Purification and characterization of the F1 ATPase from Bacillus subtilis and its uncoupler-resistant mutant ...
|
|
53004
|
Amino acid transport of y+L-type by heterodimers of 4F2hc/CD98 and members of the glycoprotein-associated ...
|
|
53005
|
Amino acid transport of y+L-type by heterodimers of 4F2hc/CD98 and members of the glycoprotein-associated ...
|
|
53006
|
Amino acid transport of y+L-type by heterodimers of 4F2hc/CD98 and members of the glycoprotein-associated ...
|
|
53007
|
Amino acid transport of y+L-type by heterodimers of 4F2hc/CD98 and members of the glycoprotein-associated ...
|
|
53008
|
Amino acid transport of y+L-type by heterodimers of 4F2hc/CD98 and members of the glycoprotein-associated ...
|
|
53009
|
Amino acid transport of y+L-type by heterodimers of 4F2hc/CD98 and members of the glycoprotein-associated ...
|
|
53010
|
Amino acid transport of y+L-type by heterodimers of 4F2hc/CD98 and members of the glycoprotein-associated ...
|
|
53011
|
Kinetic characterization of cytochrome c oxidase from Bacillus subtilis
|
|
53012
|
Kinetic characterization of cytochrome c oxidase from Bacillus subtilis
|
|
53013
|
Kinetic characterization of cytochrome c oxidase from Bacillus subtilis
|
|
53014
|
Kinetic characterization of cytochrome c oxidase from Bacillus subtilis
|
|
53015
|
Kinetic characterization of cytochrome c oxidase from Bacillus subtilis
|
|
53016
|
Kinetic characterization of cytochrome c oxidase from Bacillus subtilis
|
|
53017
|
The outB gene of Bacillus subtilis codes for NAD synthetase
|
|
53018
|
The outB gene of Bacillus subtilis codes for NAD synthetase
|
|
53019
|
5-Enolpyruvylshikimate-3-phosphate synthase of Bacillus subtilis is an allosteric enzyme. Analysis of ...
|
|
53020
|
5-Enolpyruvylshikimate-3-phosphate synthase of Bacillus subtilis is an allosteric enzyme. Analysis of ...
|
|
53021
|
5-Enolpyruvylshikimate-3-phosphate synthase of Bacillus subtilis is an allosteric enzyme. Analysis of ...
|
|
53022
|
5-Enolpyruvylshikimate-3-phosphate synthase of Bacillus subtilis is an allosteric enzyme. Analysis of ...
|
|
53023
|
5-Enolpyruvylshikimate-3-phosphate synthase of Bacillus subtilis is an allosteric enzyme. Analysis of ...
|
|
53024
|
5-Enolpyruvylshikimate-3-phosphate synthase of Bacillus subtilis is an allosteric enzyme. Analysis of ...
|
|
53025
|
5-Enolpyruvylshikimate-3-phosphate synthase of Bacillus subtilis is an allosteric enzyme. Analysis of ...
|
|
53026
|
5-Enolpyruvylshikimate-3-phosphate synthase of Bacillus subtilis is an allosteric enzyme. Analysis of ...
|
|
53027
|
5-Enolpyruvylshikimate-3-phosphate synthase of Bacillus subtilis is an allosteric enzyme. Analysis of ...
|
|
53028
|
5-Enolpyruvylshikimate-3-phosphate synthase of Bacillus subtilis is an allosteric enzyme. Analysis of ...
|
|
53029
|
5-Enolpyruvylshikimate-3-phosphate synthase of Bacillus subtilis is an allosteric enzyme. Analysis of ...
|
|
53030
|
5-Enolpyruvylshikimate-3-phosphate synthase of Bacillus subtilis is an allosteric enzyme. Analysis of ...
|
|
53031
|
5-Enolpyruvylshikimate-3-phosphate synthase of Bacillus subtilis is an allosteric enzyme. Analysis of ...
|
|
53032
|
Mechanism of mupirocin transport into sensitive and resistant bacteria
|
|
53033
|
Mechanism of mupirocin transport into sensitive and resistant bacteria
|
|
53034
|
Mechanism of mupirocin transport into sensitive and resistant bacteria
|
|
53035
|
Substrate-dependent change in the pH-activity profile of alkaline endo-1,4-beta-glucanase from an alkaline ...
|
|
53036
|
Substrate-dependent change in the pH-activity profile of alkaline endo-1,4-beta-glucanase from an alkaline ...
|
|
53037
|
Substrate-dependent change in the pH-activity profile of alkaline endo-1,4-beta-glucanase from an alkaline ...
|
|
53038
|
Substrate-dependent change in the pH-activity profile of alkaline endo-1,4-beta-glucanase from an alkaline ...
|
|
53039
|
Substrate-dependent change in the pH-activity profile of alkaline endo-1,4-beta-glucanase from an alkaline ...
|
|
53040
|
Substrate-dependent change in the pH-activity profile of alkaline endo-1,4-beta-glucanase from an alkaline ...
|
|
53041
|
Substrate-dependent change in the pH-activity profile of alkaline endo-1,4-beta-glucanase from an alkaline ...
|
|
53042
|
Substrate-dependent change in the pH-activity profile of alkaline endo-1,4-beta-glucanase from an alkaline ...
|
|
53043
|
Substrate-dependent change in the pH-activity profile of alkaline endo-1,4-beta-glucanase from an alkaline ...
|
|
53044
|
Substrate-dependent change in the pH-activity profile of alkaline endo-1,4-beta-glucanase from an alkaline ...
|
|
53045
|
Substrate-dependent change in the pH-activity profile of alkaline endo-1,4-beta-glucanase from an alkaline ...
|
|
53046
|
Substrate requirements for ErmC' methyltransferase activity
|
|
53047
|
Substrate requirements for ErmC' methyltransferase activity
|
|
53048
|
Substrate requirements for ErmC' methyltransferase activity
|
|
53049
|
Substrate requirements for ErmC' methyltransferase activity
|
|
53050
|
Ornithine transcarbamylase from Salmonella typhimurium: purification, subunit composition, kinetic analysis, ...
|
|
53051
|
Ornithine transcarbamylase from Salmonella typhimurium: purification, subunit composition, kinetic analysis, ...
|
|
53052
|
Characterization of the major citrate synthase of Bacillus subtilis
|
|
53053
|
Characterization of the major citrate synthase of Bacillus subtilis
|
|
53054
|
Specific inhibition of phosphatidate cytidylyltransferase from Bacillus subtilis membranes by cytidine ...
|
|
53055
|
Specific inhibition of phosphatidate cytidylyltransferase from Bacillus subtilis membranes by cytidine ...
|
|
53056
|
Specific inhibition of phosphatidate cytidylyltransferase from Bacillus subtilis membranes by cytidine ...
|
|
53057
|
Specific inhibition of phosphatidate cytidylyltransferase from Bacillus subtilis membranes by cytidine ...
|
|
53058
|
Specific inhibition of phosphatidate cytidylyltransferase from Bacillus subtilis membranes by cytidine ...
|
|
53059
|
Purification, characterization, and physiological function of Bacillus subtilis ornithine transcarbamylase
|
|
53060
|
Purification, characterization, and physiological function of Bacillus subtilis ornithine transcarbamylase
|
|
53061
|
Purification, characterization, and physiological function of Bacillus subtilis ornithine transcarbamylase
|
|
53062
|
Ethylene induces de novo synthesis of chlorophyllase, a chlorophyll degrading enzyme, in Citrus fruit peel
|
|
53063
|
Imidazole acetol phosphate aminotransferase in Zymomonas mobilis: molecular genetic, biochemical, and ...
|
|
53064
|
Imidazole acetol phosphate aminotransferase in Zymomonas mobilis: molecular genetic, biochemical, and ...
|
|
53065
|
Imidazole acetol phosphate aminotransferase in Zymomonas mobilis: molecular genetic, biochemical, and ...
|
|
53066
|
Molecular characterization of a phenylalanine ammonia-lyase gene (BoPAL1) from Bambusa oldhamii
|
|
53067
|
Cloning and expression of a phenylalanine ammonia-lyase gene (BoPAL2) from Bambusa oldhamii in Escherichia ...
|
|
53068
|
Cloning and expression of a phenylalanine ammonia-lyase gene (BoPAL2) from Bambusa oldhamii in Escherichia ...
|
|
53069
|
Thermolabile alanine racemase from a psychotroph, Pseudomonas fluorescens: purification and properties
|
|
53070
|
Thermolabile alanine racemase from a psychotroph, Pseudomonas fluorescens: purification and properties
|
|
53071
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53072
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53073
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53074
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53075
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53076
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53077
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53078
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53079
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53080
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53081
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53082
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53083
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53084
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53085
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53086
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53087
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53088
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53089
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53090
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53091
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53092
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53093
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53094
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53095
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53096
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53097
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53098
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53099
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53100
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53101
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53102
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53103
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53104
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53105
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53106
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53107
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53108
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53109
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53110
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53111
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53112
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53113
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53114
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53115
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53116
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53117
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53118
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53119
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53120
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53121
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53122
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53123
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53124
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53125
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53126
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53127
|
Kinetic specificities of BPN' and Carlsberg subtilisins. Mapping the aromatic binding site
|
|
53128
|
Purification and characterization of beta-D-glucosidase (beta-D-fucosidase) from Bifidobacterium breve clb ...
|
|
53129
|
Purification and characterization of beta-D-glucosidase (beta-D-fucosidase) from Bifidobacterium breve clb ...
|
|
53130
|
Characterization of pyrimidine-repressible and arginine-repressible carbamyl phosphate synthetases from ...
|
|
53131
|
Characterization of pyrimidine-repressible and arginine-repressible carbamyl phosphate synthetases from ...
|
|
53132
|
Characterization of pyrimidine-repressible and arginine-repressible carbamyl phosphate synthetases from ...
|
|
53133
|
Characterization of pyrimidine-repressible and arginine-repressible carbamyl phosphate synthetases from ...
|
|
53134
|
Characterization of pyrimidine-repressible and arginine-repressible carbamyl phosphate synthetases from ...
|
|
53135
|
Characterization of pyrimidine-repressible and arginine-repressible carbamyl phosphate synthetases from ...
|
|
53136
|
Characterization of the surfactin synthetase multi-enzyme complex
|
|
53137
|
Characterization of the surfactin synthetase multi-enzyme complex
|
|
53138
|
Characterization of the surfactin synthetase multi-enzyme complex
|
|
53139
|
3-Carboxy-cis,cis-muconate lactonizing enzyme from Pseudomonas putida is homologous to the class II fumarase ...
|
|
53140
|
3-Carboxy-cis,cis-muconate lactonizing enzyme from Pseudomonas putida is homologous to the class II fumarase ...
|
|
53141
|
3-Carboxy-cis,cis-muconate lactonizing enzyme from Pseudomonas putida is homologous to the class II fumarase ...
|
|
53142
|
Regulation of UDP-glucose pyrophosphorylase isozyme UGP5 associated with cold-sweetening resistance in ...
|
|
53143
|
Regulation of UDP-glucose pyrophosphorylase isozyme UGP5 associated with cold-sweetening resistance in ...
|
|
53144
|
Regulation of UDP-glucose pyrophosphorylase isozyme UGP5 associated with cold-sweetening resistance in ...
|
|
53145
|
Regulation of UDP-glucose pyrophosphorylase isozyme UGP5 associated with cold-sweetening resistance in ...
|
|
53146
|
Differential metabolism of 4-n- and 4-tert-octylphenols in perfused rat liver
|
|
53147
|
Differential metabolism of 4-n- and 4-tert-octylphenols in perfused rat liver
|
|
53148
|
Differential metabolism of 4-n- and 4-tert-octylphenols in perfused rat liver
|
|
53149
|
Differential metabolism of 4-n- and 4-tert-octylphenols in perfused rat liver
|
|
53150
|
Differential metabolism of 4-n- and 4-tert-octylphenols in perfused rat liver
|
|
53151
|
Differential metabolism of 4-n- and 4-tert-octylphenols in perfused rat liver
|
|
53152
|
Differential metabolism of 4-n- and 4-tert-octylphenols in perfused rat liver
|
|
53153
|
Differential metabolism of 4-n- and 4-tert-octylphenols in perfused rat liver
|
|
53154
|
Differential metabolism of 4-n- and 4-tert-octylphenols in perfused rat liver
|
|
53155
|
Differential metabolism of 4-n- and 4-tert-octylphenols in perfused rat liver
|
|
53156
|
Differential metabolism of 4-n- and 4-tert-octylphenols in perfused rat liver
|
|
53157
|
Differential metabolism of 4-n- and 4-tert-octylphenols in perfused rat liver
|
|
53158
|
Biosynthesis of riboflavin. Lumazine synthase of Escherichia coli
|
|
53159
|
Biosynthesis of riboflavin. Lumazine synthase of Escherichia coli
|
|
53160
|
Purification, kinetic and thermodynamic characterization of soluble acid invertase from sugarcane (Saccharum ...
|
|
53161
|
GC-MS assay for hepatic DDAH activity in diabetic and non-diabetic rats by measuring dimethylamine (DMA) ...
|
|
53162
|
GC-MS assay for hepatic DDAH activity in diabetic and non-diabetic rats by measuring dimethylamine (DMA) ...
|
|
53163
|
GC-MS assay for hepatic DDAH activity in diabetic and non-diabetic rats by measuring dimethylamine (DMA) ...
|
|
53164
|
GC-MS assay for hepatic DDAH activity in diabetic and non-diabetic rats by measuring dimethylamine (DMA) ...
|
|
53165
|
GC-MS assay for hepatic DDAH activity in diabetic and non-diabetic rats by measuring dimethylamine (DMA) ...
|
|
53166
|
Subtilisin from Bacillus subtilis strain 72. The influence of substrate structure, temperature and pH on ...
|
|
53167
|
Subtilisin from Bacillus subtilis strain 72. The influence of substrate structure, temperature and pH on ...
|
|
53168
|
Subtilisin from Bacillus subtilis strain 72. The influence of substrate structure, temperature and pH on ...
|
|
53169
|
Subtilisin from Bacillus subtilis strain 72. The influence of substrate structure, temperature and pH on ...
|
|
53170
|
Subtilisin from Bacillus subtilis strain 72. The influence of substrate structure, temperature and pH on ...
|
|
53171
|
Subtilisin from Bacillus subtilis strain 72. The influence of substrate structure, temperature and pH on ...
|
|
53172
|
Subtilisin from Bacillus subtilis strain 72. The influence of substrate structure, temperature and pH on ...
|
|
53173
|
Subtilisin from Bacillus subtilis strain 72. The influence of substrate structure, temperature and pH on ...
|
|
53174
|
Subtilisin from Bacillus subtilis strain 72. The influence of substrate structure, temperature and pH on ...
|
|
53175
|
Subtilisin from Bacillus subtilis strain 72. The influence of substrate structure, temperature and pH on ...
|
|
53176
|
Subtilisin from Bacillus subtilis strain 72. The influence of substrate structure, temperature and pH on ...
|
|
53177
|
Subtilisin from Bacillus subtilis strain 72. The influence of substrate structure, temperature and pH on ...
|
|
53178
|
Subtilisin from Bacillus subtilis strain 72. The influence of substrate structure, temperature and pH on ...
|
|
53179
|
Subtilisin from Bacillus subtilis strain 72. The influence of substrate structure, temperature and pH on ...
|
|
53180
|
Subtilisin from Bacillus subtilis strain 72. The influence of substrate structure, temperature and pH on ...
|
|
53181
|
Subtilisin from Bacillus subtilis strain 72. The influence of substrate structure, temperature and pH on ...
|
|
53182
|
Histidine ammonia-lyase from Streptomyces griseus
|
|
53183
|
Histidine ammonia-lyase from Streptomyces griseus
|
|
53184
|
Histidine ammonia-lyase from Streptomyces griseus
|
|
53185
|
Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants--structural ...
|
|
53186
|
Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants--structural ...
|
|
53187
|
Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants--structural ...
|
|
53188
|
Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants--structural ...
|
|
53189
|
Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants--structural ...
|
|
53190
|
Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants--structural ...
|
|
53191
|
Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants--structural ...
|
|
53192
|
Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants--structural ...
|
|
53193
|
Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants--structural ...
|
|
53194
|
Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants--structural ...
|
|
53195
|
Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants--structural ...
|
|
53196
|
Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants--structural ...
|
|
53197
|
Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants--structural ...
|
|
53198
|
Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants--structural ...
|
|
53199
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53200
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53201
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53202
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53203
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53204
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53205
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53206
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53207
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53208
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53209
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53210
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53211
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53212
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53213
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53214
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53215
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53216
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53217
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53218
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53219
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53220
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53221
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53222
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53223
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53224
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53225
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53226
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53227
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53228
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53229
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53230
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53231
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53232
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53233
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53234
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53235
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53236
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53237
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53238
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53239
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53240
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53241
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53242
|
The role of lysine-234 in beta-lactamase catalysis probed by site-directed mutagenesis
|
|
53243
|
Partial purification and characterisation of sucrose synthase in sugarcane
|
|
53244
|
Partial purification and characterisation of sucrose synthase in sugarcane
|
|
53245
|
Partial purification and characterisation of sucrose synthase in sugarcane
|
|
53246
|
Partial purification and characterisation of sucrose synthase in sugarcane
|
|
53247
|
Partial purification and characterisation of sucrose synthase in sugarcane
|
|
53248
|
Partial purification and characterisation of sucrose synthase in sugarcane
|
|
53249
|
Partial purification and characterisation of sucrose synthase in sugarcane
|
|
53250
|
Partial purification and characterisation of sucrose synthase in sugarcane
|
|
53251
|
Partial purification and characterisation of sucrose synthase in sugarcane
|
|
53252
|
Partial purification and characterisation of sucrose synthase in sugarcane
|
|
53253
|
Partial purification and characterisation of sucrose synthase in sugarcane
|
|
53254
|
Partial purification and characterisation of sucrose synthase in sugarcane
|
|
53255
|
Molecular cloning, expression and characterization of hyoscyamine 6beta-hydroxylase from hairy roots of ...
|
|
53256
|
Molecular cloning, expression and characterization of hyoscyamine 6beta-hydroxylase from hairy roots of ...
|
|
53257
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53258
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53259
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53260
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53261
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53262
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53263
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53264
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53265
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53266
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53267
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53268
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53269
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53270
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53271
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53272
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53273
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53274
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53275
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53276
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53277
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53278
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53279
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53280
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53281
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53282
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53283
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53284
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53285
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53286
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53287
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53288
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53289
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53290
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53291
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53292
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53293
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53294
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53295
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53296
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53297
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53298
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53299
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53300
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53301
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53302
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53303
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53304
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53305
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53306
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53307
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53308
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53309
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53310
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53311
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53312
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53313
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53314
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53315
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53316
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53317
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53318
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53319
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53320
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53321
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53322
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53323
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53324
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53325
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53326
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53327
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53328
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53329
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53330
|
The dopamine D1D receptor. Cloning and characterization of three pharmacologically distinct D1-like receptors ...
|
|
53331
|
Putrescine N-methyltransferases--a structure-function analysis
|
|
53332
|
Putrescine N-methyltransferases--a structure-function analysis
|
|
53333
|
Putrescine N-methyltransferases--a structure-function analysis
|
|
53334
|
Putrescine N-methyltransferases--a structure-function analysis
|
|
53335
|
Putrescine N-methyltransferases--a structure-function analysis
|
|
53336
|
Putrescine N-methyltransferases--a structure-function analysis
|
|
53337
|
Putrescine N-methyltransferases--a structure-function analysis
|
|
53338
|
Putrescine N-methyltransferases--a structure-function analysis
|
|
53339
|
Putrescine N-methyltransferases--a structure-function analysis
|
|
53340
|
De novo purine nucleotide biosynthesis: cloning, sequencing and expression of a chicken PurH cDNA encoding ...
|
|
53341
|
De novo purine nucleotide biosynthesis: cloning, sequencing and expression of a chicken PurH cDNA encoding ...
|
|
53342
|
Structure-assisted redesign of a protein-zinc-binding site with femtomolar affinity
|
|
53343
|
Structure-assisted redesign of a protein-zinc-binding site with femtomolar affinity
|
|
53344
|
Structure-assisted redesign of a protein-zinc-binding site with femtomolar affinity
|
|
53345
|
Structure-assisted redesign of a protein-zinc-binding site with femtomolar affinity
|
|
53346
|
Structure-assisted redesign of a protein-zinc-binding site with femtomolar affinity
|
|
53347
|
Structure-assisted redesign of a protein-zinc-binding site with femtomolar affinity
|
|
53348
|
Structure-assisted redesign of a protein-zinc-binding site with femtomolar affinity
|
|
53349
|
Neuraminidase assay in cultured human fibroblasts: in situ versus in vitro procedures
|
|
53350
|
Biotin synthase: purification, characterization as a [2Fe-2S]cluster protein, and in vitro activity of the ...
|
|
53351
|
Biotin synthase: purification, characterization as a [2Fe-2S]cluster protein, and in vitro activity of the ...
|
|
53352
|
Biotin synthase: purification, characterization as a [2Fe-2S]cluster protein, and in vitro activity of the ...
|
|
53353
|
Biotin synthase: purification, characterization as a [2Fe-2S]cluster protein, and in vitro activity of the ...
|
|
53354
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53355
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53356
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53357
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53358
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53359
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53360
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53361
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53362
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53363
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53364
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53365
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53366
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53367
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53368
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53369
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53370
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53371
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53372
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53373
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53374
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53375
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53376
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53377
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53378
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53379
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53380
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53381
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53382
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53383
|
Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 ...
|
|
53384
|
Cumulative stabilizing effects of glycine to alanine substitutions in Bacillus subtilis neutral protease
|
|
53385
|
Cumulative stabilizing effects of glycine to alanine substitutions in Bacillus subtilis neutral protease
|
|
53386
|
Cumulative stabilizing effects of glycine to alanine substitutions in Bacillus subtilis neutral protease
|
|
53387
|
Cumulative stabilizing effects of glycine to alanine substitutions in Bacillus subtilis neutral protease
|
|
53388
|
Cumulative stabilizing effects of glycine to alanine substitutions in Bacillus subtilis neutral protease
|
|
53389
|
Cumulative stabilizing effects of glycine to alanine substitutions in Bacillus subtilis neutral protease
|
|
53390
|
Cumulative stabilizing effects of glycine to alanine substitutions in Bacillus subtilis neutral protease
|
|
53391
|
Cumulative stabilizing effects of glycine to alanine substitutions in Bacillus subtilis neutral protease
|
|
53392
|
Cumulative stabilizing effects of glycine to alanine substitutions in Bacillus subtilis neutral protease
|
|
53393
|
Cumulative stabilizing effects of glycine to alanine substitutions in Bacillus subtilis neutral protease
|
|
53394
|
Cumulative stabilizing effects of glycine to alanine substitutions in Bacillus subtilis neutral protease
|
|
53395
|
Cumulative stabilizing effects of glycine to alanine substitutions in Bacillus subtilis neutral protease
|
|
53396
|
Cumulative stabilizing effects of glycine to alanine substitutions in Bacillus subtilis neutral protease
|
|
53397
|
Cumulative stabilizing effects of glycine to alanine substitutions in Bacillus subtilis neutral protease
|
|
53398
|
Cumulative stabilizing effects of glycine to alanine substitutions in Bacillus subtilis neutral protease
|
|
53399
|
Cumulative stabilizing effects of glycine to alanine substitutions in Bacillus subtilis neutral protease
|
|
53400
|
Acyl amino acid derivatives as novel inhibitors of influenza neuraminidase
|
|
53401
|
Acyl amino acid derivatives as novel inhibitors of influenza neuraminidase
|
|
53402
|
Acyl amino acid derivatives as novel inhibitors of influenza neuraminidase
|
|
53403
|
Acyl amino acid derivatives as novel inhibitors of influenza neuraminidase
|
|
53404
|
Glutamate synthase from Bacillus subtilis PCI 219
|
|
53405
|
Glutamate synthase from Bacillus subtilis PCI 219
|
|
53406
|
Glutamate synthase from Bacillus subtilis PCI 219
|
|
53407
|
Glutamate synthase from Bacillus subtilis PCI 219
|
|
53408
|
Glutamate synthase from Bacillus subtilis PCI 219
|
|
53409
|
Glutamate synthase from Bacillus subtilis PCI 219
|
|
53410
|
Glutamate synthase from Bacillus subtilis PCI 219
|
|
53411
|
Glutamate synthase from Bacillus subtilis PCI 219
|
|
53412
|
Glutamate synthase from Bacillus subtilis PCI 219
|
|
53413
|
Glutamate synthase from Bacillus subtilis PCI 219
|
|
53414
|
Glutamate synthase from Bacillus subtilis PCI 219
|
|
53415
|
Glutamate synthase from Bacillus subtilis PCI 219
|
|
53416
|
Glutamate synthase from Bacillus subtilis PCI 219
|
|
53417
|
Glutamate synthase from Bacillus subtilis PCI 219
|
|
53418
|
Glutamate synthase from Bacillus subtilis PCI 219
|
|
53419
|
Glutamate synthase from Bacillus subtilis PCI 219
|
|
53420
|
Glutamate synthase from Bacillus subtilis PCI 219
|
|
53421
|
Glutamate synthase from Bacillus subtilis PCI 219
|
|
53422
|
Glutamate synthase from Bacillus subtilis PCI 219
|
|
53423
|
Glutamate synthase from Bacillus subtilis PCI 219
|
|
53424
|
Glutamate synthase from Bacillus subtilis PCI 219
|
|
53425
|
Glutamate synthase from Bacillus subtilis PCI 219
|
|
53426
|
Glutamate synthase from Bacillus subtilis PCI 219
|
|
53427
|
Glutamate synthase from Bacillus subtilis PCI 219
|
|
53428
|
Glutamate synthase from Bacillus subtilis PCI 219
|
|
53429
|
Enzymic mechanism of starch synthesis in ripening rice grains. 3. Mechanism of the sucrose-starch conversion.
|
|
53430
|
Enzymic mechanism of starch synthesis in ripening rice grains. 3. Mechanism of the sucrose-starch conversion.
|
|
53431
|
N-ethoxycarbonyl-D-phenylalanyl-L-prolyl-alpha-azalysine p-nitrophenyl ester: a novel, high selective and ...
|
|
53432
|
N-ethoxycarbonyl-D-phenylalanyl-L-prolyl-alpha-azalysine p-nitrophenyl ester: a novel, high selective and ...
|
|
53433
|
N-ethoxycarbonyl-D-phenylalanyl-L-prolyl-alpha-azalysine p-nitrophenyl ester: a novel, high selective and ...
|
|
53434
|
Identification of domains involved in tetramerization and malate inhibition of maize C4-NADP-malic enzyme
|
|
53435
|
Identification of domains involved in tetramerization and malate inhibition of maize C4-NADP-malic enzyme
|
|
53436
|
Identification of domains involved in tetramerization and malate inhibition of maize C4-NADP-malic enzyme
|
|
53437
|
Identification of domains involved in tetramerization and malate inhibition of maize C4-NADP-malic enzyme
|
|
53438
|
Identification of domains involved in tetramerization and malate inhibition of maize C4-NADP-malic enzyme
|
|
53439
|
Identification of domains involved in tetramerization and malate inhibition of maize C4-NADP-malic enzyme
|
|
53440
|
Identification of domains involved in tetramerization and malate inhibition of maize C4-NADP-malic enzyme
|
|
53441
|
Identification of domains involved in tetramerization and malate inhibition of maize C4-NADP-malic enzyme
|
|
53442
|
Identification of domains involved in tetramerization and malate inhibition of maize C4-NADP-malic enzyme
|
|
53443
|
Identification of domains involved in tetramerization and malate inhibition of maize C4-NADP-malic enzyme
|
|
53444
|
Identification of domains involved in tetramerization and malate inhibition of maize C4-NADP-malic enzyme
|
|
53445
|
Identification of domains involved in tetramerization and malate inhibition of maize C4-NADP-malic enzyme
|
|
53446
|
Identification of domains involved in tetramerization and malate inhibition of maize C4-NADP-malic enzyme
|
|
53447
|
Identification of domains involved in tetramerization and malate inhibition of maize C4-NADP-malic enzyme
|
|
53448
|
Identification of domains involved in tetramerization and malate inhibition of maize C4-NADP-malic enzyme
|
|
53449
|
Identification of domains involved in tetramerization and malate inhibition of maize C4-NADP-malic enzyme
|
|
53450
|
Identification of domains involved in tetramerization and malate inhibition of maize C4-NADP-malic enzyme
|
|
53451
|
Identification of domains involved in tetramerization and malate inhibition of maize C4-NADP-malic enzyme
|
|
53452
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53453
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53454
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53455
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53456
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53457
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53458
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53459
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53460
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53461
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53462
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53463
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53464
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53465
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53466
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53467
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53468
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53469
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53470
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53471
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53472
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53473
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53474
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53475
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53476
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53477
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53478
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53479
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53480
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53481
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53482
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53483
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53484
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53485
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53486
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53487
|
Two glutamine synthetases from Bacillus caldolyticus, an extreme thermophile. Isolation, physicochemical and ...
|
|
53488
|
The phosphoenolpyruvate : methyl-alpha-d-glucoside phosphotransferase system in Bacillus subtilis Marburg : ...
|
|
53489
|
Expression, purification and characterization of recombinant sucrose synthase 1 from Solanum tuberosum L. for ...
|
|
53490
|
Expression, purification and characterization of recombinant sucrose synthase 1 from Solanum tuberosum L. for ...
|
|
53491
|
Expression, purification and characterization of recombinant sucrose synthase 1 from Solanum tuberosum L. for ...
|
|
53492
|
Expression, purification and characterization of recombinant sucrose synthase 1 from Solanum tuberosum L. for ...
|
|
53493
|
Expression, purification and characterization of recombinant sucrose synthase 1 from Solanum tuberosum L. for ...
|
|
53494
|
Expression, purification and characterization of recombinant sucrose synthase 1 from Solanum tuberosum L. for ...
|
|
53495
|
Expression, purification and characterization of recombinant sucrose synthase 1 from Solanum tuberosum L. for ...
|
|
53496
|
Expression, purification and characterization of recombinant sucrose synthase 1 from Solanum tuberosum L. for ...
|
|
53497
|
Expression, purification and characterization of recombinant sucrose synthase 1 from Solanum tuberosum L. for ...
|
|
53498
|
Expression, purification and characterization of recombinant sucrose synthase 1 from Solanum tuberosum L. for ...
|
|
53499
|
Expression, purification and characterization of recombinant sucrose synthase 1 from Solanum tuberosum L. for ...
|
|
53500
|
The Donor Substrate Spectrum of Recombinant Sucrose Synthase 1 from Potato for the Synthesis of Sucrose ...
|
|
53501
|
The Donor Substrate Spectrum of Recombinant Sucrose Synthase 1 from Potato for the Synthesis of Sucrose ...
|
|
53502
|
Identification and expression of NEU3, a novel human sialidase associated to the plasma membrane
|
|
53503
|
Identification and expression of NEU3, a novel human sialidase associated to the plasma membrane
|
|
53504
|
Identification and expression of NEU3, a novel human sialidase associated to the plasma membrane
|
|
53505
|
Identification and expression of NEU3, a novel human sialidase associated to the plasma membrane
|
|
53506
|
Identification and expression of NEU3, a novel human sialidase associated to the plasma membrane
|
|
53507
|
Identification and expression of NEU3, a novel human sialidase associated to the plasma membrane
|
|
53508
|
Identification and expression of NEU3, a novel human sialidase associated to the plasma membrane
|
|
53509
|
Identification and expression of NEU3, a novel human sialidase associated to the plasma membrane
|
|
53510
|
Identification and expression of NEU3, a novel human sialidase associated to the plasma membrane
|
|
53511
|
Characterization of Recombinant Sucrose Synthase 1 from Potato for the Synthesis of Sucrose Analogues
|
|
53512
|
Characterization of Recombinant Sucrose Synthase 1 from Potato for the Synthesis of Sucrose Analogues
|
|
53513
|
Characterization of Recombinant Sucrose Synthase 1 from Potato for the Synthesis of Sucrose Analogues
|
|
53514
|
Characterization of Recombinant Sucrose Synthase 1 from Potato for the Synthesis of Sucrose Analogues
|
|
53515
|
Characterization of Recombinant Sucrose Synthase 1 from Potato for the Synthesis of Sucrose Analogues
|
|
53516
|
Characterization of Recombinant Sucrose Synthase 1 from Potato for the Synthesis of Sucrose Analogues
|
|
53517
|
Severe X-linked mitochondrial encephalomyopathy associated with a mutation in apoptosis-inducing factor.
|
|
53518
|
Severe X-linked mitochondrial encephalomyopathy associated with a mutation in apoptosis-inducing factor.
|
|
53519
|
Purification of 6-phosphogluconate dehydrogenase from parsley (Petroselinum hortense) leaves and investigation ...
|
|
53520
|
Purification of 6-phosphogluconate dehydrogenase from parsley (Petroselinum hortense) leaves and investigation ...
|
|
53521
|
Molecular and biochemical characterization of cytosolic phosphoglucomutase in wheat endosperm (Triticum ...
|
|
53522
|
Molecular and biochemical characterization of cytosolic phosphoglucomutase in wheat endosperm (Triticum ...
|
|
53523
|
Arabidopsis mitochondria have two basic amino acid transporters with partially overlapping specificities and ...
|
|
53524
|
Arabidopsis mitochondria have two basic amino acid transporters with partially overlapping specificities and ...
|
|
53525
|
Photoaffinity labeling of a cell surface polyamine binding protein
|
|
53526
|
Photoaffinity labeling of a cell surface polyamine binding protein
|
|
53527
|
Photoaffinity labeling of a cell surface polyamine binding protein
|
|
53528
|
Photoaffinity labeling of a cell surface polyamine binding protein
|
|
53529
|
Photoaffinity labeling of a cell surface polyamine binding protein
|
|
53530
|
Regulation of Polyamine Transport by Polyamines and Polyamine Analogs
|
|
53531
|
Regulation of Polyamine Transport by Polyamines and Polyamine Analogs
|
|
53532
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53533
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53534
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53535
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53536
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53537
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53538
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53539
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53540
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53541
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53542
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53543
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53544
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53545
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53546
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53547
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53548
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53549
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53550
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53551
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53552
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53553
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53554
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53555
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53556
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53557
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53558
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53559
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53560
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53561
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53562
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53563
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53564
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53565
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53566
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53567
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53568
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53569
|
The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct ...
|
|
53570
|
Crystal structures of human Delta4-3-ketosteroid 5beta-reductase (AKR1D1) reveal the presence of an ...
|
|
53571
|
Crystal structures of human Delta4-3-ketosteroid 5beta-reductase (AKR1D1) reveal the presence of an ...
|
|
53572
|
Crystal structures of human Delta4-3-ketosteroid 5beta-reductase (AKR1D1) reveal the presence of an ...
|
|
53573
|
Crystal structures of human Delta4-3-ketosteroid 5beta-reductase (AKR1D1) reveal the presence of an ...
|
|
53574
|
Characterization of the human placental membrane receptor for transcobalamin II-cobalamin
|
|
53575
|
Characterization of the human placental membrane receptor for transcobalamin II-cobalamin
|
|
53576
|
Characterization of the human placental membrane receptor for transcobalamin II-cobalamin
|
|
53577
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53578
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53579
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53580
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53581
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53582
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53583
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53584
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53585
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53586
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53587
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53588
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53589
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53590
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53591
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53592
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53593
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53594
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53595
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53596
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53597
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53598
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53599
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53600
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53601
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53602
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53603
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53604
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53605
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53606
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53607
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53608
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53609
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53610
|
Inhibitors of phenylalanine ammonia-lyase: 1-aminobenzylphosphonic acids substituted in the benzene ring.
|
|
53611
|
A simple and specific assay of glycosyltransferase and glycosidase activities by an enzyme-linked ...
|
|
53612
|
A simple and specific assay of glycosyltransferase and glycosidase activities by an enzyme-linked ...
|
|
53613
|
A simple and specific assay of glycosyltransferase and glycosidase activities by an enzyme-linked ...
|
|
53614
|
A simple and specific assay of glycosyltransferase and glycosidase activities by an enzyme-linked ...
|
|
53615
|
A simple and specific assay of glycosyltransferase and glycosidase activities by an enzyme-linked ...
|
|
53616
|
A simple and specific assay of glycosyltransferase and glycosidase activities by an enzyme-linked ...
|
|
53617
|
A simple and specific assay of glycosyltransferase and glycosidase activities by an enzyme-linked ...
|
|
53618
|
A peptidase in human platelets that deamidates tachykinins. Probable identity with the lysosomal "protective ...
|
|
53619
|
A peptidase in human platelets that deamidates tachykinins. Probable identity with the lysosomal "protective ...
|
|
53620
|
A peptidase in human platelets that deamidates tachykinins. Probable identity with the lysosomal "protective ...
|
|
53621
|
Oligomerization status, with the monomer as active species, defines catalytic efficiency of UDP-glucose ...
|
|
53622
|
Oligomerization status, with the monomer as active species, defines catalytic efficiency of UDP-glucose ...
|
|
53623
|
Oligomerization status, with the monomer as active species, defines catalytic efficiency of UDP-glucose ...
|
|
53624
|
Oligomerization status, with the monomer as active species, defines catalytic efficiency of UDP-glucose ...
|
|
53625
|
Metabolic engineering in yeast demonstrates that S-adenosylmethionine controls flux through the ...
|
|
53626
|
Metabolic engineering in yeast demonstrates that S-adenosylmethionine controls flux through the ...
|
|
53627
|
Metabolic engineering in yeast demonstrates that S-adenosylmethionine controls flux through the ...
|
|
53628
|
Metabolic engineering in yeast demonstrates that S-adenosylmethionine controls flux through the ...
|
|
53629
|
Metabolic engineering in yeast demonstrates that S-adenosylmethionine controls flux through the ...
|
|
53630
|
Metabolic engineering in yeast demonstrates that S-adenosylmethionine controls flux through the ...
|
|
53631
|
Metabolic engineering in yeast demonstrates that S-adenosylmethionine controls flux through the ...
|
|
53632
|
Metabolic engineering in yeast demonstrates that S-adenosylmethionine controls flux through the ...
|
|
53633
|
Glucuronidation of the environmental oestrogen bisphenol A by an isoform of UDP-glucuronosyltransferase, ...
|
|
53634
|
Glucuronidation of the environmental oestrogen bisphenol A by an isoform of UDP-glucuronosyltransferase, ...
|
|
53635
|
Glucuronidation of the environmental oestrogen bisphenol A by an isoform of UDP-glucuronosyltransferase, ...
|
|
53636
|
Glucuronidation of the environmental oestrogen bisphenol A by an isoform of UDP-glucuronosyltransferase, ...
|
|
53637
|
Glucuronidation of the environmental oestrogen bisphenol A by an isoform of UDP-glucuronosyltransferase, ...
|
|
53638
|
Glucuronidation of the environmental oestrogen bisphenol A by an isoform of UDP-glucuronosyltransferase, ...
|
|
53639
|
Conversion of human steroid 5β-reductase (AKR1D1) into 3β-hydroxysteroid dehydrogenase by single point ...
|
|
53640
|
Conversion of human steroid 5β-reductase (AKR1D1) into 3β-hydroxysteroid dehydrogenase by single point ...
|
|
53641
|
Conversion of human steroid 5β-reductase (AKR1D1) into 3β-hydroxysteroid dehydrogenase by single point ...
|
|
53642
|
Conversion of human steroid 5β-reductase (AKR1D1) into 3β-hydroxysteroid dehydrogenase by single point ...
|
|
53643
|
Conversion of human steroid 5β-reductase (AKR1D1) into 3β-hydroxysteroid dehydrogenase by single point ...
|
|
53644
|
Conversion of human steroid 5β-reductase (AKR1D1) into 3β-hydroxysteroid dehydrogenase by single point ...
|
|
53645
|
Conversion of human steroid 5β-reductase (AKR1D1) into 3β-hydroxysteroid dehydrogenase by single point ...
|
|
53646
|
Conversion of human steroid 5β-reductase (AKR1D1) into 3β-hydroxysteroid dehydrogenase by single point ...
|
|
53647
|
Conversion of human steroid 5β-reductase (AKR1D1) into 3β-hydroxysteroid dehydrogenase by single point ...
|
|
53648
|
Conversion of human steroid 5β-reductase (AKR1D1) into 3β-hydroxysteroid dehydrogenase by single point ...
|
|
53649
|
Conversion of human steroid 5β-reductase (AKR1D1) into 3β-hydroxysteroid dehydrogenase by single point ...
|
|
53650
|
Conversion of human steroid 5β-reductase (AKR1D1) into 3β-hydroxysteroid dehydrogenase by single point ...
|
|
53651
|
Conversion of human steroid 5β-reductase (AKR1D1) into 3β-hydroxysteroid dehydrogenase by single point ...
|
|
53652
|
Conversion of human steroid 5β-reductase (AKR1D1) into 3β-hydroxysteroid dehydrogenase by single point ...
|
|
53653
|
Conversion of human steroid 5β-reductase (AKR1D1) into 3β-hydroxysteroid dehydrogenase by single point ...
|
|
53654
|
Conversion of human steroid 5β-reductase (AKR1D1) into 3β-hydroxysteroid dehydrogenase by single point ...
|
|
53655
|
Conversion of human steroid 5β-reductase (AKR1D1) into 3β-hydroxysteroid dehydrogenase by single point ...
|
|
53656
|
Conversion of human steroid 5β-reductase (AKR1D1) into 3β-hydroxysteroid dehydrogenase by single point ...
|
|
53657
|
Conversion of human steroid 5β-reductase (AKR1D1) into 3β-hydroxysteroid dehydrogenase by single point ...
|
|
53658
|
Conversion of human steroid 5β-reductase (AKR1D1) into 3β-hydroxysteroid dehydrogenase by single point ...
|
|
53659
|
Anaerobic metabolism of 3-hydroxybenzoate by the denitrifying bacterium Thauera aromatica
|
|
53660
|
Anaerobic metabolism of 3-hydroxybenzoate by the denitrifying bacterium Thauera aromatica
|
|
53661
|
Anaerobic metabolism of 3-hydroxybenzoate by the denitrifying bacterium Thauera aromatica
|
|
53662
|
Anaerobic metabolism of 3-hydroxybenzoate by the denitrifying bacterium Thauera aromatica
|
|
53663
|
Overproduction, purification, and characterization of recombinant aspartate semialdehyde dehydrogenase from ...
|
|
53664
|
Overproduction, purification, and characterization of recombinant aspartate semialdehyde dehydrogenase from ...
|
|
53665
|
The Arabidopsis thaliana trp5 mutant has a feedback-resistant anthranilate synthase and elevated soluble ...
|
|
53666
|
The Arabidopsis thaliana trp5 mutant has a feedback-resistant anthranilate synthase and elevated soluble ...
|
|
53667
|
The Arabidopsis thaliana trp5 mutant has a feedback-resistant anthranilate synthase and elevated soluble ...
|
|
53668
|
The Arabidopsis thaliana trp5 mutant has a feedback-resistant anthranilate synthase and elevated soluble ...
|
|
53669
|
Cloning and molecular genetic characterization of the Escherichia coli gntR, gntK, and gntU genes of GntI, the ...
|
|
53670
|
Cloning and molecular genetic characterization of the Escherichia coli gntR, gntK, and gntU genes of GntI, the ...
|
|
53671
|
Direct evidence for nitric oxide production by a nitric-oxide synthase-like protein from Bacillus subtilis
|
|
53672
|
Direct evidence for nitric oxide production by a nitric-oxide synthase-like protein from Bacillus subtilis
|
|
53673
|
Purification and characterization of a recombinant pea cytoplasmic fructose-1,6-bisphosphatase
|
|
53674
|
Purification and characterization of a recombinant pea cytoplasmic fructose-1,6-bisphosphatase
|
|
53675
|
Purification and characterization of a recombinant pea cytoplasmic fructose-1,6-bisphosphatase
|
|
53676
|
Purification and characterization of a recombinant pea cytoplasmic fructose-1,6-bisphosphatase
|
|
53677
|
Purification and characterization of a recombinant pea cytoplasmic fructose-1,6-bisphosphatase
|
|
53678
|
Purification and characterization of a recombinant pea cytoplasmic fructose-1,6-bisphosphatase
|
|
53679
|
Rainbow trout (Oncorhynchus mykiss) cystatin C: expression in Escherichia coli and properties of the ...
|
|
53680
|
The dimer contact area of sorghum NADP-malate dehydrogenase: role of aspartate 101 in dimer stability and ...
|
|
53681
|
The dimer contact area of sorghum NADP-malate dehydrogenase: role of aspartate 101 in dimer stability and ...
|
|
53682
|
The dimer contact area of sorghum NADP-malate dehydrogenase: role of aspartate 101 in dimer stability and ...
|
|
53683
|
Subcellular localization of ADPglucose pyrophosphorylase in developing wheat endosperm and analysis of the ...
|
|
53684
|
Subcellular localization of ADPglucose pyrophosphorylase in developing wheat endosperm and analysis of the ...
|
|
53685
|
Subcellular localization of ADPglucose pyrophosphorylase in developing wheat endosperm and analysis of the ...
|
|
53686
|
Subcellular localization of ADPglucose pyrophosphorylase in developing wheat endosperm and analysis of the ...
|
|
53687
|
Subcellular localization of ADPglucose pyrophosphorylase in developing wheat endosperm and analysis of the ...
|
|
53688
|
Subcellular localization of ADPglucose pyrophosphorylase in developing wheat endosperm and analysis of the ...
|
|
53689
|
Subcellular localization of ADPglucose pyrophosphorylase in developing wheat endosperm and analysis of the ...
|
|
53690
|
Subcellular localization of ADPglucose pyrophosphorylase in developing wheat endosperm and analysis of the ...
|
|
53691
|
Cloning, expression, and characterization of a root-form phosphoenolpyruvate carboxylase from Zea mays: ...
|
|
53692
|
Cloning, expression, and characterization of a root-form phosphoenolpyruvate carboxylase from Zea mays: ...
|
|
53693
|
Cloning, expression, and characterization of a root-form phosphoenolpyruvate carboxylase from Zea mays: ...
|
|
53694
|
Cloning, expression, and characterization of a root-form phosphoenolpyruvate carboxylase from Zea mays: ...
|
|
53695
|
Cloning, expression, and characterization of a root-form phosphoenolpyruvate carboxylase from Zea mays: ...
|
|
53696
|
Cloning, expression, and characterization of a root-form phosphoenolpyruvate carboxylase from Zea mays: ...
|
|
53697
|
Cloning, expression, and characterization of a root-form phosphoenolpyruvate carboxylase from Zea mays: ...
|
|
53698
|
Cloning, expression, and characterization of a root-form phosphoenolpyruvate carboxylase from Zea mays: ...
|
|
53699
|
Cloning, expression, and characterization of a root-form phosphoenolpyruvate carboxylase from Zea mays: ...
|
|
53700
|
Cloning, expression, and characterization of a root-form phosphoenolpyruvate carboxylase from Zea mays: ...
|
|
53701
|
Cloning, expression, and characterization of a root-form phosphoenolpyruvate carboxylase from Zea mays: ...
|
|
53702
|
Cloning, expression, and characterization of a root-form phosphoenolpyruvate carboxylase from Zea mays: ...
|
|
53703
|
Peptidoglycan N-acetylglucosamine deacetylases from Bacillus cereus, highly conserved proteins in Bacillus ...
|
|
53704
|
Peptidoglycan N-acetylglucosamine deacetylases from Bacillus cereus, highly conserved proteins in Bacillus ...
|
|
53705
|
Peptidoglycan N-acetylglucosamine deacetylases from Bacillus cereus, highly conserved proteins in Bacillus ...
|
|
53706
|
Peptidoglycan N-acetylglucosamine deacetylases from Bacillus cereus, highly conserved proteins in Bacillus ...
|
|
53707
|
Peptidoglycan N-acetylglucosamine deacetylases from Bacillus cereus, highly conserved proteins in Bacillus ...
|
|
53708
|
Peptidoglycan N-acetylglucosamine deacetylases from Bacillus cereus, highly conserved proteins in Bacillus ...
|
|
53709
|
Peptidoglycan N-acetylglucosamine deacetylases from Bacillus cereus, highly conserved proteins in Bacillus ...
|
|
53710
|
Peptidoglycan N-acetylglucosamine deacetylases from Bacillus cereus, highly conserved proteins in Bacillus ...
|
|
53711
|
Peptidoglycan N-acetylglucosamine deacetylases from Bacillus cereus, highly conserved proteins in Bacillus ...
|
|
53712
|
Peptidoglycan N-acetylglucosamine deacetylases from Bacillus cereus, highly conserved proteins in Bacillus ...
|
|
53713
|
Identification, purification, and molecular cloning of a putative plastidic glucose translocator
|
|
53714
|
Sulfation of sialyl N-acetyllactosamine oligosaccharides and fetuin oligosaccharides by keratan sulfate ...
|
|
53715
|
Sulfation of sialyl N-acetyllactosamine oligosaccharides and fetuin oligosaccharides by keratan sulfate ...
|
|
53716
|
Sulfation of sialyl N-acetyllactosamine oligosaccharides and fetuin oligosaccharides by keratan sulfate ...
|
|
53717
|
Sulfation of sialyl N-acetyllactosamine oligosaccharides and fetuin oligosaccharides by keratan sulfate ...
|
|
53718
|
Characterization of aspartate aminotransferase isoenzymes from leaves of Lupinus albus L. cv Estoril
|
|
53719
|
Characterization of aspartate aminotransferase isoenzymes from leaves of Lupinus albus L. cv Estoril
|
|
53720
|
Characterization of aspartate aminotransferase isoenzymes from leaves of Lupinus albus L. cv Estoril
|
|
53721
|
Characterization of aspartate aminotransferase isoenzymes from leaves of Lupinus albus L. cv Estoril
|
|
53722
|
Characterization of aspartate aminotransferase isoenzymes from leaves of Lupinus albus L. cv Estoril
|
|
53723
|
Characterization of aspartate aminotransferase isoenzymes from leaves of Lupinus albus L. cv Estoril
|
|
53724
|
Characterization of aspartate aminotransferase isoenzymes from leaves of Lupinus albus L. cv Estoril
|
|
53725
|
Characterization of aspartate aminotransferase isoenzymes from leaves of Lupinus albus L. cv Estoril
|
|
53726
|
Characterization of aspartate aminotransferase isoenzymes from leaves of Lupinus albus L. cv Estoril
|
|
53727
|
Characterization of aspartate aminotransferase isoenzymes from leaves of Lupinus albus L. cv Estoril
|
|
53728
|
Characterization of aspartate aminotransferase isoenzymes from leaves of Lupinus albus L. cv Estoril
|
|
53729
|
Characterization of aspartate aminotransferase isoenzymes from leaves of Lupinus albus L. cv Estoril
|
|
53730
|
Characterization of aspartate aminotransferase isoenzymes from leaves of Lupinus albus L. cv Estoril
|
|
53731
|
Characterization of aspartate aminotransferase isoenzymes from leaves of Lupinus albus L. cv Estoril
|
|
53732
|
Characterization of aspartate aminotransferase isoenzymes from leaves of Lupinus albus L. cv Estoril
|
|
53733
|
Characterization of aspartate aminotransferase isoenzymes from leaves of Lupinus albus L. cv Estoril
|
|
53734
|
Characterization of aspartate aminotransferase isoenzymes from leaves of Lupinus albus L. cv Estoril
|
|
53735
|
Characterization of aspartate aminotransferase isoenzymes from leaves of Lupinus albus L. cv Estoril
|
|
53736
|
Characterization of aspartate aminotransferase isoenzymes from leaves of Lupinus albus L. cv Estoril
|
|
53737
|
Characterization of aspartate aminotransferase isoenzymes from leaves of Lupinus albus L. cv Estoril
|
|
53738
|
Maize C4 and non-C4 NADP-dependent malic enzymes are encoded by distinct genes derived from a ...
|
|
53739
|
Maize C4 and non-C4 NADP-dependent malic enzymes are encoded by distinct genes derived from a ...
|
|
53740
|
Probing the inhibitor selectivity pocket of human 20α-hydroxysteroid dehydrogenase (AKR1C1) with X-ray ...
|
|
53741
|
Probing the inhibitor selectivity pocket of human 20α-hydroxysteroid dehydrogenase (AKR1C1) with X-ray ...
|
|
53742
|
Probing the inhibitor selectivity pocket of human 20α-hydroxysteroid dehydrogenase (AKR1C1) with X-ray ...
|
|
53743
|
Probing the inhibitor selectivity pocket of human 20α-hydroxysteroid dehydrogenase (AKR1C1) with X-ray ...
|
|
53744
|
Probing the inhibitor selectivity pocket of human 20α-hydroxysteroid dehydrogenase (AKR1C1) with X-ray ...
|
|
53745
|
Probing the inhibitor selectivity pocket of human 20α-hydroxysteroid dehydrogenase (AKR1C1) with X-ray ...
|
|
53746
|
Probing the inhibitor selectivity pocket of human 20α-hydroxysteroid dehydrogenase (AKR1C1) with X-ray ...
|
|
53747
|
Probing the inhibitor selectivity pocket of human 20α-hydroxysteroid dehydrogenase (AKR1C1) with X-ray ...
|
|
53748
|
The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2
|
|
53749
|
The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2
|
|
53750
|
The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2
|
|
53751
|
The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2
|
|
53752
|
The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2
|
|
53753
|
The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2
|
|
53754
|
The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2
|
|
53755
|
The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2
|
|
53756
|
The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2
|
|
53757
|
The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2
|
|
53758
|
The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2
|
|
53759
|
The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2
|
|
53760
|
The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2
|
|
53761
|
The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2
|
|
53762
|
The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2
|
|
53763
|
Isolation and enzymic properties of levansucrase secreted by Acetobacter diazotrophicus SRT4, a bacterium ...
|
|
53764
|
Purification of tomato sucrose synthase phosphorylated isoforms by Fe(III)-immobilized metal affinity ...
|
|
53765
|
Purification of tomato sucrose synthase phosphorylated isoforms by Fe(III)-immobilized metal affinity ...
|
|
53766
|
Purification of tomato sucrose synthase phosphorylated isoforms by Fe(III)-immobilized metal affinity ...
|
|
53767
|
Purification of tomato sucrose synthase phosphorylated isoforms by Fe(III)-immobilized metal affinity ...
|
|
53768
|
Cloning, sequencing, and expression of a beta-amylase gene from Bacillus cereus var. mycoides and ...
|
|
53769
|
Cloning, sequencing, and expression of a beta-amylase gene from Bacillus cereus var. mycoides and ...
|
|
53770
|
Cloning, sequencing, and expression of a beta-amylase gene from Bacillus cereus var. mycoides and ...
|
|
53771
|
Purification and kinetic properties of UDP-glucose dehydrogenase from sugarcane
|
|
53772
|
Purification and kinetic properties of UDP-glucose dehydrogenase from sugarcane
|
|
53773
|
Purification and kinetic properties of UDP-glucose dehydrogenase from sugarcane
|
|
53774
|
Purification and kinetic properties of UDP-glucose dehydrogenase from sugarcane
|
|
53775
|
Substrate specificity, membrane topology, and activity regulation of human alkaline ceramidase 2 (ACER2).
|
|
53776
|
Substrate specificity, membrane topology, and activity regulation of human alkaline ceramidase 2 (ACER2).
|
|
53777
|
Substrate specificity, membrane topology, and activity regulation of human alkaline ceramidase 2 (ACER2).
|
|
53778
|
Substrate specificity, membrane topology, and activity regulation of human alkaline ceramidase 2 (ACER2).
|
|
53779
|
Substrate specificity, membrane topology, and activity regulation of human alkaline ceramidase 2 (ACER2).
|
|
53780
|
Substrate specificity, membrane topology, and activity regulation of human alkaline ceramidase 2 (ACER2).
|
|
53781
|
Substrate specificity, membrane topology, and activity regulation of human alkaline ceramidase 2 (ACER2).
|
|
53782
|
Substrate specificity, membrane topology, and activity regulation of human alkaline ceramidase 2 (ACER2).
|
|
53783
|
Substrate specificity, membrane topology, and activity regulation of human alkaline ceramidase 2 (ACER2).
|
|
53784
|
Substrate specificity, membrane topology, and activity regulation of human alkaline ceramidase 2 (ACER2).
|
|
53785
|
Peptide specificity and lipid activation of the lysosomal transport complex ABCB9 (TAPL)
|
|
53786
|
Effects of 5' leader and 3' trailer structures on pre-tRNA processing by nuclear RNase P
|
|
53787
|
Effects of 5' leader and 3' trailer structures on pre-tRNA processing by nuclear RNase P
|
|
53788
|
Effects of 5' leader and 3' trailer structures on pre-tRNA processing by nuclear RNase P
|
|
53789
|
Effects of 5' leader and 3' trailer structures on pre-tRNA processing by nuclear RNase P
|
|
53790
|
Effects of 5' leader and 3' trailer structures on pre-tRNA processing by nuclear RNase P
|
|
53791
|
Effects of 5' leader and 3' trailer structures on pre-tRNA processing by nuclear RNase P
|
|
53792
|
Effects of 5' leader and 3' trailer structures on pre-tRNA processing by nuclear RNase P
|
|
53793
|
Effects of 5' leader and 3' trailer structures on pre-tRNA processing by nuclear RNase P
|
|
53794
|
Effects of 5' leader and 3' trailer structures on pre-tRNA processing by nuclear RNase P
|
|
53795
|
Effect of desialylation on the biological properties of human plasminogen
|
|
53796
|
Effect of desialylation on the biological properties of human plasminogen
|
|
53797
|
Molecular and biochemical characterization of an Arabidopsis thaliana arogenate dehydrogenase with two highly ...
|
|
53798
|
Molecular and biochemical characterization of an Arabidopsis thaliana arogenate dehydrogenase with two highly ...
|
|
53799
|
Molecular and biochemical characterization of an Arabidopsis thaliana arogenate dehydrogenase with two highly ...
|
|
53800
|
Selective and ATP-dependent Translocation of Peptides by the Homodimeric ATP Binding Cassette Transporter ...
|
|
53801
|
Selective and ATP-dependent Translocation of Peptides by the Homodimeric ATP Binding Cassette Transporter ...
|
|
53802
|
Selective and ATP-dependent Translocation of Peptides by the Homodimeric ATP Binding Cassette Transporter ...
|
|
53803
|
Selective and ATP-dependent Translocation of Peptides by the Homodimeric ATP Binding Cassette Transporter ...
|
|
53804
|
Selective and ATP-dependent Translocation of Peptides by the Homodimeric ATP Binding Cassette Transporter ...
|
|
53805
|
Inhibition of p-hydroxyphenylpyruvate dioxygenase by the diketonitrile of isoxaflutole: a case of half-site ...
|
|
53806
|
Identification of the maize amyloplast stromal 112-kD protein as a plastidic starch phosphorylase
|
|
53807
|
Identification of the maize amyloplast stromal 112-kD protein as a plastidic starch phosphorylase
|
|
53808
|
Two essential regions for tRNA recognition in Bacillus subtilis tryptophanyl-tRNA synthetase
|
|
53809
|
Two essential regions for tRNA recognition in Bacillus subtilis tryptophanyl-tRNA synthetase
|
|
53810
|
Two essential regions for tRNA recognition in Bacillus subtilis tryptophanyl-tRNA synthetase
|
|
53811
|
Two essential regions for tRNA recognition in Bacillus subtilis tryptophanyl-tRNA synthetase
|
|
53812
|
Two essential regions for tRNA recognition in Bacillus subtilis tryptophanyl-tRNA synthetase
|
|
53813
|
Two essential regions for tRNA recognition in Bacillus subtilis tryptophanyl-tRNA synthetase
|
|
53814
|
Two essential regions for tRNA recognition in Bacillus subtilis tryptophanyl-tRNA synthetase
|
|
53815
|
Cloning and characterization of the arginine-specific carbamoyl-phosphate synthetase from Bacillus ...
|
|
53816
|
Cloning and characterization of the arginine-specific carbamoyl-phosphate synthetase from Bacillus ...
|
|
53817
|
Cloning and characterization of the arginine-specific carbamoyl-phosphate synthetase from Bacillus ...
|
|
53818
|
Cloning and characterization of the arginine-specific carbamoyl-phosphate synthetase from Bacillus ...
|
|
53819
|
Cloning and characterization of the arginine-specific carbamoyl-phosphate synthetase from Bacillus ...
|
|
53820
|
Cloning and characterization of the arginine-specific carbamoyl-phosphate synthetase from Bacillus ...
|
|
53821
|
Cloning and characterization of the arginine-specific carbamoyl-phosphate synthetase from Bacillus ...
|
|
53822
|
Cloning and characterization of the arginine-specific carbamoyl-phosphate synthetase from Bacillus ...
|
|
53823
|
Cloning and characterization of the arginine-specific carbamoyl-phosphate synthetase from Bacillus ...
|
|
53824
|
Purification and characterization of recombinant, human acid ceramidase. Catalytic reactions and interactions ...
|
|
53825
|
Purification and characterization of recombinant, human acid ceramidase. Catalytic reactions and interactions ...
|
|
53826
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53827
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53828
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53829
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53830
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53831
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53832
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53833
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53834
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53835
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53836
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53837
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53838
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53839
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53840
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53841
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53842
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53843
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53844
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53845
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53846
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53847
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53848
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53849
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53850
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53851
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53852
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53853
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53854
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53855
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53856
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53857
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53858
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53859
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53860
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53861
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53862
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53863
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53864
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53865
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53866
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53867
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53868
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53869
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53870
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53871
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53872
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53873
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53874
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53875
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53876
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53877
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53878
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53879
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53880
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53881
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53882
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53883
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53884
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53885
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53886
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53887
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53888
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53889
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53890
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53891
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53892
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53893
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53894
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53895
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53896
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53897
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53898
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53899
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53900
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53901
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53902
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53903
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53904
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53905
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53906
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53907
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53908
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53909
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53910
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53911
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53912
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53913
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53914
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53915
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53916
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53917
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53918
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53919
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53920
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53921
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53922
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53923
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53924
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53925
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53926
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53927
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53928
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53929
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53930
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53931
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53932
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53933
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53934
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53935
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53936
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53937
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53938
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53939
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53940
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53941
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53942
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53943
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53944
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53945
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53946
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53947
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53948
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53949
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53950
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53951
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53952
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53953
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53954
|
Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H- ...
|
|
53955
|
Urea is a product of ureidoglycolate degradation in chickpea. Purification and characterization of the ...
|
|
53956
|
Urea is a product of ureidoglycolate degradation in chickpea. Purification and characterization of the ...
|
|
53957
|
Purification and characterization of two sucrose synthase isoforms from Japanese pear fruit.
|
|
53958
|
Purification and characterization of two sucrose synthase isoforms from Japanese pear fruit.
|
|
53959
|
Purification and characterization of two sucrose synthase isoforms from Japanese pear fruit.
|
|
53960
|
Purification and characterization of two sucrose synthase isoforms from Japanese pear fruit.
|
|
53961
|
Purification and characterization of two sucrose synthase isoforms from Japanese pear fruit.
|
|
53962
|
Purification and characterization of two sucrose synthase isoforms from Japanese pear fruit.
|
|
53963
|
Purification and characterization of two sucrose synthase isoforms from Japanese pear fruit.
|
|
53964
|
Purification and characterization of two sucrose synthase isoforms from Japanese pear fruit.
|
|
53965
|
Purification and characterization of two sucrose synthase isoforms from Japanese pear fruit.
|
|
53966
|
Purification and characterization of two sucrose synthase isoforms from Japanese pear fruit.
|
|
53967
|
Purification and characterization of two sucrose synthase isoforms from Japanese pear fruit.
|
|
53968
|
Purification and characterization of two sucrose synthase isoforms from Japanese pear fruit.
|
|
53969
|
Kinetic and mutational analyses of the regulation of phosphoribulokinase by thioredoxins.
|
|
53970
|
Kinetic and mutational analyses of the regulation of phosphoribulokinase by thioredoxins.
|
|
53971
|
Kinetic and mutational analyses of the regulation of phosphoribulokinase by thioredoxins.
|
|
53972
|
Kinetic and mutational analyses of the regulation of phosphoribulokinase by thioredoxins.
|
|
53973
|
Kinetic and mutational analyses of the regulation of phosphoribulokinase by thioredoxins.
|
|
53974
|
Kinetic and mutational analyses of the regulation of phosphoribulokinase by thioredoxins.
|
|
53975
|
Kinetic and mutational analyses of the regulation of phosphoribulokinase by thioredoxins.
|
|
53976
|
Kinetic and mutational analyses of the regulation of phosphoribulokinase by thioredoxins.
|
|
53977
|
Kinetic and mutational analyses of the regulation of phosphoribulokinase by thioredoxins.
|
|
53978
|
Kinetic and mutational analyses of the regulation of phosphoribulokinase by thioredoxins.
|
|
53979
|
Kinetic and mutational analyses of the regulation of phosphoribulokinase by thioredoxins.
|
|
53980
|
Characterization of the surfactin synthetase C-terminal thioesterase domain as a cyclic depsipeptide synthase
|
|
53981
|
Characterization of the surfactin synthetase C-terminal thioesterase domain as a cyclic depsipeptide synthase
|
|
53982
|
Characterization of the surfactin synthetase C-terminal thioesterase domain as a cyclic depsipeptide synthase
|
|
53983
|
Characterization of the surfactin synthetase C-terminal thioesterase domain as a cyclic depsipeptide synthase
|
|
53984
|
Characterization of the surfactin synthetase C-terminal thioesterase domain as a cyclic depsipeptide synthase
|
|
53985
|
Characterization of the surfactin synthetase C-terminal thioesterase domain as a cyclic depsipeptide synthase
|
|
53986
|
Characterization of the surfactin synthetase C-terminal thioesterase domain as a cyclic depsipeptide synthase
|
|
53987
|
Characterization of the surfactin synthetase C-terminal thioesterase domain as a cyclic depsipeptide synthase
|
|
53988
|
Characterization of the surfactin synthetase C-terminal thioesterase domain as a cyclic depsipeptide synthase
|
|
53989
|
Characterization of the surfactin synthetase C-terminal thioesterase domain as a cyclic depsipeptide synthase
|
|
53990
|
Characterization of the surfactin synthetase C-terminal thioesterase domain as a cyclic depsipeptide synthase
|
|
53991
|
Characterization of the surfactin synthetase C-terminal thioesterase domain as a cyclic depsipeptide synthase
|
|
53992
|
Characterization of the surfactin synthetase C-terminal thioesterase domain as a cyclic depsipeptide synthase
|
|
53993
|
Characterization of the surfactin synthetase C-terminal thioesterase domain as a cyclic depsipeptide synthase
|
|
53994
|
Characterization of the surfactin synthetase C-terminal thioesterase domain as a cyclic depsipeptide synthase
|
|
53995
|
Characterization of the surfactin synthetase C-terminal thioesterase domain as a cyclic depsipeptide synthase
|
|
53996
|
Characterization of the surfactin synthetase C-terminal thioesterase domain as a cyclic depsipeptide synthase
|
|
53997
|
Characterization of the surfactin synthetase C-terminal thioesterase domain as a cyclic depsipeptide synthase
|
|
53998
|
Characterization of the surfactin synthetase C-terminal thioesterase domain as a cyclic depsipeptide synthase
|
|
53999
|
Characterization of the surfactin synthetase C-terminal thioesterase domain as a cyclic depsipeptide synthase
|
|
54000
|
Characterization of the surfactin synthetase C-terminal thioesterase domain as a cyclic depsipeptide synthase
|





 If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.
If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.