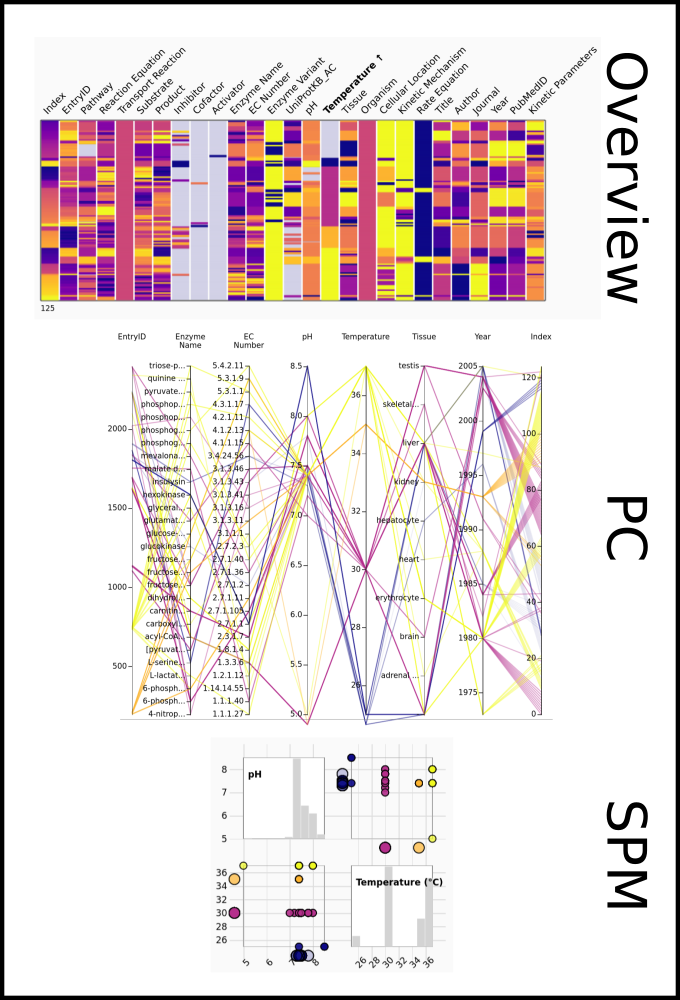
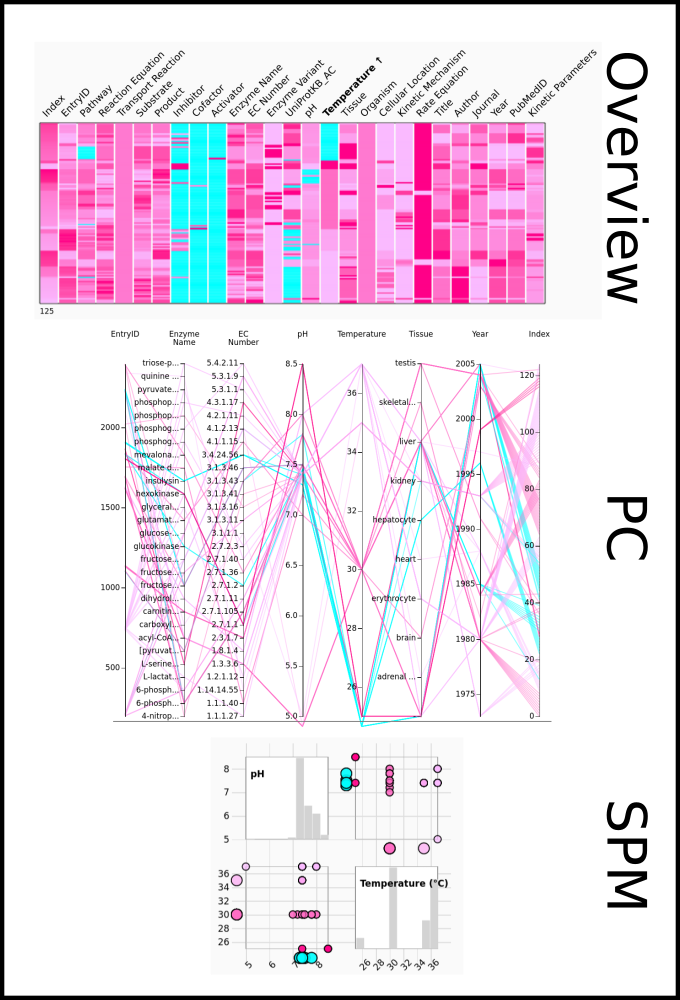
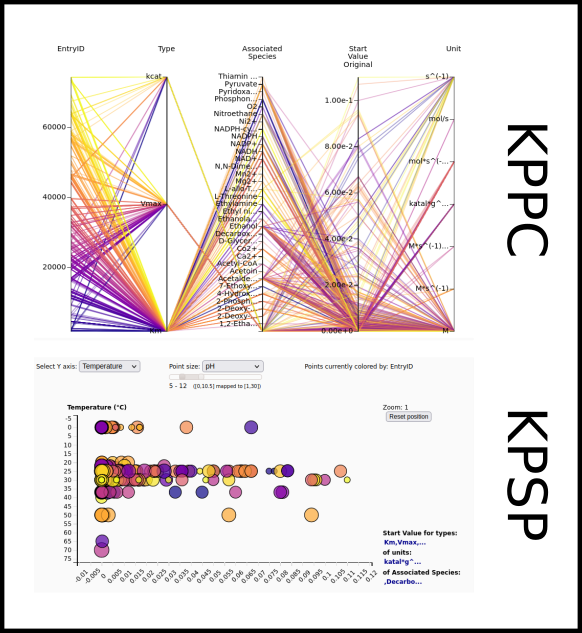
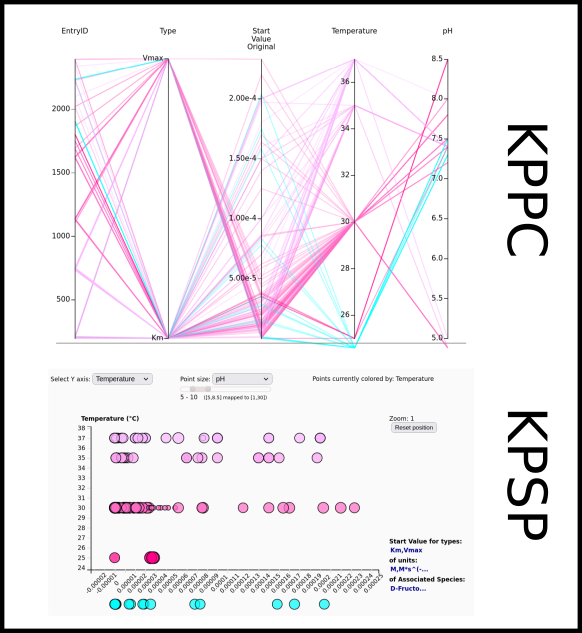
To improve SABIO-RK data interoperability, semantic markup was added to web pages as described and defined by
This structured information makes it easier to discover, collate and analyse our data.
|
52001
|
Glycine oxidase from Bacillus subtilis: role of histidine 244 and methionine 261
|
|
52002
|
Glycine oxidase from Bacillus subtilis: role of histidine 244 and methionine 261
|
|
52003
|
Glycine oxidase from Bacillus subtilis: role of histidine 244 and methionine 261
|
|
52004
|
Glycine oxidase from Bacillus subtilis: role of histidine 244 and methionine 261
|
|
52005
|
Glycine oxidase from Bacillus subtilis: role of histidine 244 and methionine 261
|
|
52006
|
Glycine oxidase from Bacillus subtilis: role of histidine 244 and methionine 261
|
|
52007
|
Glycine oxidase from Bacillus subtilis: role of histidine 244 and methionine 261
|
|
52008
|
Glycine oxidase from Bacillus subtilis: role of histidine 244 and methionine 261
|
|
52009
|
Purification and enzymic properties of the fructosyltransferase of Streptococcus salivarius ATCC 25975
|
|
52010
|
Engineering tropane biosynthetic pathway in Hyoscyamus niger hairy root cultures
|
|
52011
|
Engineering tropane biosynthetic pathway in Hyoscyamus niger hairy root cultures
|
|
52012
|
Engineering tropane biosynthetic pathway in Hyoscyamus niger hairy root cultures
|
|
52013
|
Engineering tropane biosynthetic pathway in Hyoscyamus niger hairy root cultures
|
|
52014
|
CYP79B1 from Sinapis alba converts tryptophan to indole-3-acetaldoxime
|
|
52015
|
Identification and optimization of tyrosine hydroxylase activity in Mucuna pruriens DC. var. utilis
|
|
52016
|
Identification and optimization of tyrosine hydroxylase activity in Mucuna pruriens DC. var. utilis
|
|
52017
|
Identification and optimization of tyrosine hydroxylase activity in Mucuna pruriens DC. var. utilis
|
|
52018
|
Thermoanaerobacterium thermosulfurigenes cyclodextrin glycosyltransferase
|
|
52019
|
Thermoanaerobacterium thermosulfurigenes cyclodextrin glycosyltransferase
|
|
52020
|
Thermoanaerobacterium thermosulfurigenes cyclodextrin glycosyltransferase
|
|
52021
|
Thermoanaerobacterium thermosulfurigenes cyclodextrin glycosyltransferase
|
|
52022
|
Thermoanaerobacterium thermosulfurigenes cyclodextrin glycosyltransferase
|
|
52023
|
Thermoanaerobacterium thermosulfurigenes cyclodextrin glycosyltransferase
|
|
52024
|
Thermoanaerobacterium thermosulfurigenes cyclodextrin glycosyltransferase
|
|
52025
|
Thermoanaerobacterium thermosulfurigenes cyclodextrin glycosyltransferase
|
|
52026
|
Thermoanaerobacterium thermosulfurigenes cyclodextrin glycosyltransferase
|
|
52027
|
Thermoanaerobacterium thermosulfurigenes cyclodextrin glycosyltransferase
|
|
52028
|
Thermoanaerobacterium thermosulfurigenes cyclodextrin glycosyltransferase
|
|
52029
|
Thermoanaerobacterium thermosulfurigenes cyclodextrin glycosyltransferase
|
|
52030
|
Thermoanaerobacterium thermosulfurigenes cyclodextrin glycosyltransferase
|
|
52031
|
Thermoanaerobacterium thermosulfurigenes cyclodextrin glycosyltransferase
|
|
52032
|
Kinetic studies of the uracil phosphoribosyltransferase reaction catalyzed by the Bacillus subtilis pyrimidine ...
|
|
52033
|
Kinetic studies of the uracil phosphoribosyltransferase reaction catalyzed by the Bacillus subtilis pyrimidine ...
|
|
52034
|
Kinetic studies of the uracil phosphoribosyltransferase reaction catalyzed by the Bacillus subtilis pyrimidine ...
|
|
52035
|
Kinetic studies of the uracil phosphoribosyltransferase reaction catalyzed by the Bacillus subtilis pyrimidine ...
|
|
52036
|
Kinetic studies of the uracil phosphoribosyltransferase reaction catalyzed by the Bacillus subtilis pyrimidine ...
|
|
52037
|
Kinetic studies of the uracil phosphoribosyltransferase reaction catalyzed by the Bacillus subtilis pyrimidine ...
|
|
52038
|
Kinetic studies of the uracil phosphoribosyltransferase reaction catalyzed by the Bacillus subtilis pyrimidine ...
|
|
52039
|
Kinetic studies of the uracil phosphoribosyltransferase reaction catalyzed by the Bacillus subtilis pyrimidine ...
|
|
52040
|
Kinetic studies of the uracil phosphoribosyltransferase reaction catalyzed by the Bacillus subtilis pyrimidine ...
|
|
52041
|
Kinetic studies of the uracil phosphoribosyltransferase reaction catalyzed by the Bacillus subtilis pyrimidine ...
|
|
52042
|
Kinetic studies of the uracil phosphoribosyltransferase reaction catalyzed by the Bacillus subtilis pyrimidine ...
|
|
52043
|
Kinetic studies of the uracil phosphoribosyltransferase reaction catalyzed by the Bacillus subtilis pyrimidine ...
|
|
52044
|
Overexpression, purification and characterization of Mycobacterium bovis BCG alcohol dehydrogenase
|
|
52045
|
Overexpression, purification and characterization of Mycobacterium bovis BCG alcohol dehydrogenase
|
|
52046
|
Overexpression, purification and characterization of Mycobacterium bovis BCG alcohol dehydrogenase
|
|
52047
|
Overexpression, purification and characterization of Mycobacterium bovis BCG alcohol dehydrogenase
|
|
52048
|
Overexpression, purification and characterization of Mycobacterium bovis BCG alcohol dehydrogenase
|
|
52049
|
Overexpression, purification and characterization of Mycobacterium bovis BCG alcohol dehydrogenase
|
|
52050
|
Overexpression, purification and characterization of Mycobacterium bovis BCG alcohol dehydrogenase
|
|
52051
|
Overexpression, purification and characterization of Mycobacterium bovis BCG alcohol dehydrogenase
|
|
52052
|
Overexpression, purification and characterization of Mycobacterium bovis BCG alcohol dehydrogenase
|
|
52053
|
Overexpression, purification and characterization of Mycobacterium bovis BCG alcohol dehydrogenase
|
|
52054
|
YjeQ, an essential, conserved, uncharacterized protein from Escherichia coli, is an unusual GTPase with ...
|
|
52055
|
YjeQ, an essential, conserved, uncharacterized protein from Escherichia coli, is an unusual GTPase with ...
|
|
52056
|
YjeQ, an essential, conserved, uncharacterized protein from Escherichia coli, is an unusual GTPase with ...
|
|
52057
|
YjeQ, an essential, conserved, uncharacterized protein from Escherichia coli, is an unusual GTPase with ...
|
|
52058
|
YjeQ, an essential, conserved, uncharacterized protein from Escherichia coli, is an unusual GTPase with ...
|
|
52059
|
YjeQ, an essential, conserved, uncharacterized protein from Escherichia coli, is an unusual GTPase with ...
|
|
52060
|
YjeQ, an essential, conserved, uncharacterized protein from Escherichia coli, is an unusual GTPase with ...
|
|
52061
|
YjeQ, an essential, conserved, uncharacterized protein from Escherichia coli, is an unusual GTPase with ...
|
|
52062
|
A point mutation of valine-311 to methionine in Bacillus subtilis protoporphyrinogen oxidase does not greatly ...
|
|
52063
|
A point mutation of valine-311 to methionine in Bacillus subtilis protoporphyrinogen oxidase does not greatly ...
|
|
52064
|
A point mutation of valine-311 to methionine in Bacillus subtilis protoporphyrinogen oxidase does not greatly ...
|
|
52065
|
A point mutation of valine-311 to methionine in Bacillus subtilis protoporphyrinogen oxidase does not greatly ...
|
|
52066
|
Purification and Properties of Adenosine 5'-Triphosphate-D-Glucose 6-Phosphotransferase from Rat Liver
|
|
52067
|
Purification and Properties of Adenosine 5'-Triphosphate-D-Glucose 6-Phosphotransferase from Rat Liver
|
|
52068
|
Purification and Properties of Adenosine 5'-Triphosphate-D-Glucose 6-Phosphotransferase from Rat Liver
|
|
52069
|
Purification and Properties of Adenosine 5'-Triphosphate-D-Glucose 6-Phosphotransferase from Rat Liver
|
|
52070
|
Purification and Properties of Adenosine 5'-Triphosphate-D-Glucose 6-Phosphotransferase from Rat Liver
|
|
52071
|
Purification and Properties of Adenosine 5'-Triphosphate-D-Glucose 6-Phosphotransferase from Rat Liver
|
|
52072
|
Purification and Properties of Adenosine 5'-Triphosphate-D-Glucose 6-Phosphotransferase from Rat Liver
|
|
52073
|
Purification and Properties of Adenosine 5'-Triphosphate-D-Glucose 6-Phosphotransferase from Rat Liver
|
|
52074
|
Purification and Properties of Adenosine 5'-Triphosphate-D-Glucose 6-Phosphotransferase from Rat Liver
|
|
52075
|
Purification and Properties of Adenosine 5'-Triphosphate-D-Glucose 6-Phosphotransferase from Rat Liver
|
|
52076
|
Purification and Properties of Adenosine 5'-Triphosphate-D-Glucose 6-Phosphotransferase from Rat Liver
|
|
52077
|
Purification and Properties of Adenosine 5'-Triphosphate-D-Glucose 6-Phosphotransferase from Rat Liver
|
|
52078
|
Purification and Properties of Adenosine 5'-Triphosphate-D-Glucose 6-Phosphotransferase from Rat Liver
|
|
52079
|
Purification and Properties of Adenosine 5'-Triphosphate-D-Glucose 6-Phosphotransferase from Rat Liver
|
|
52080
|
Purification and Properties of Adenosine 5'-Triphosphate-D-Glucose 6-Phosphotransferase from Rat Liver
|
|
52081
|
Purification and Properties of Adenosine 5'-Triphosphate-D-Glucose 6-Phosphotransferase from Rat Liver
|
|
52082
|
Purification and Properties of Adenosine 5'-Triphosphate-D-Glucose 6-Phosphotransferase from Rat Liver
|
|
52083
|
Purification and Properties of Adenosine 5'-Triphosphate-D-Glucose 6-Phosphotransferase from Rat Liver
|
|
52084
|
Purification and Properties of Adenosine 5'-Triphosphate-D-Glucose 6-Phosphotransferase from Rat Liver
|
|
52085
|
An extremely thermostable aldolase from Sulfolobus solfataricus with specificity for non-phosphorylated ...
|
|
52086
|
An extremely thermostable aldolase from Sulfolobus solfataricus with specificity for non-phosphorylated ...
|
|
52087
|
An extremely thermostable aldolase from Sulfolobus solfataricus with specificity for non-phosphorylated ...
|
|
52088
|
An extremely thermostable aldolase from Sulfolobus solfataricus with specificity for non-phosphorylated ...
|
|
52089
|
An extremely thermostable aldolase from Sulfolobus solfataricus with specificity for non-phosphorylated ...
|
|
52090
|
An extremely thermostable aldolase from Sulfolobus solfataricus with specificity for non-phosphorylated ...
|
|
52091
|
An extremely thermostable aldolase from Sulfolobus solfataricus with specificity for non-phosphorylated ...
|
|
52092
|
Induction of serotonin biosynthesis is uncoupled from the coordinated induction of tryptophan biosynthesis in ...
|
|
52093
|
Purification and characterization of two soluble acid invertase isozymes from Japanese pear fruit
|
|
52094
|
Purification and characterization of two soluble acid invertase isozymes from Japanese pear fruit
|
|
52095
|
Folate synthesis in plants: the first step of the pterin branch is mediated by a unique bimodular GTP ...
|
|
52096
|
Purification and biochemical characterization of the ErmSF macrolide-lincosamide-streptogramin B resistance ...
|
|
52097
|
Purification and biochemical characterization of the ErmSF macrolide-lincosamide-streptogramin B resistance ...
|
|
52098
|
Purification and biochemical characterization of the ErmSF macrolide-lincosamide-streptogramin B resistance ...
|
|
52099
|
Purification and biochemical characterization of the ErmSF macrolide-lincosamide-streptogramin B resistance ...
|
|
52100
|
Arabidopsis phosphomannose isomerase 1, but not phosphomannose isomerase 2, is essential for ascorbic acid ...
|
|
52101
|
Arabidopsis phosphomannose isomerase 1, but not phosphomannose isomerase 2, is essential for ascorbic acid ...
|
|
52102
|
Arabidopsis phosphomannose isomerase 1, but not phosphomannose isomerase 2, is essential for ascorbic acid ...
|
|
52103
|
Arabidopsis phosphomannose isomerase 1, but not phosphomannose isomerase 2, is essential for ascorbic acid ...
|
|
52104
|
Arabidopsis phosphomannose isomerase 1, but not phosphomannose isomerase 2, is essential for ascorbic acid ...
|
|
52105
|
Arabidopsis phosphomannose isomerase 1, but not phosphomannose isomerase 2, is essential for ascorbic acid ...
|
|
52106
|
Arabidopsis phosphomannose isomerase 1, but not phosphomannose isomerase 2, is essential for ascorbic acid ...
|
|
52107
|
Arabidopsis phosphomannose isomerase 1, but not phosphomannose isomerase 2, is essential for ascorbic acid ...
|
|
52108
|
Purification, kinetic properties, and intracellular concentration of SpoIIE, an integral membrane protein that ...
|
|
52109
|
Random mutagenesis into the conserved Gly154 of subtilisin E: isolation and characterization of the revertant ...
|
|
52110
|
Random mutagenesis into the conserved Gly154 of subtilisin E: isolation and characterization of the revertant ...
|
|
52111
|
Random mutagenesis into the conserved Gly154 of subtilisin E: isolation and characterization of the revertant ...
|
|
52112
|
Random mutagenesis into the conserved Gly154 of subtilisin E: isolation and characterization of the revertant ...
|
|
52113
|
Random mutagenesis into the conserved Gly154 of subtilisin E: isolation and characterization of the revertant ...
|
|
52114
|
Random mutagenesis into the conserved Gly154 of subtilisin E: isolation and characterization of the revertant ...
|
|
52115
|
Random mutagenesis into the conserved Gly154 of subtilisin E: isolation and characterization of the revertant ...
|
|
52116
|
Random mutagenesis into the conserved Gly154 of subtilisin E: isolation and characterization of the revertant ...
|
|
52117
|
Random mutagenesis into the conserved Gly154 of subtilisin E: isolation and characterization of the revertant ...
|
|
52118
|
Random mutagenesis into the conserved Gly154 of subtilisin E: isolation and characterization of the revertant ...
|
|
52119
|
Random mutagenesis into the conserved Gly154 of subtilisin E: isolation and characterization of the revertant ...
|
|
52120
|
Random mutagenesis into the conserved Gly154 of subtilisin E: isolation and characterization of the revertant ...
|
|
52121
|
Random mutagenesis into the conserved Gly154 of subtilisin E: isolation and characterization of the revertant ...
|
|
52122
|
Random mutagenesis into the conserved Gly154 of subtilisin E: isolation and characterization of the revertant ...
|
|
52123
|
Random mutagenesis into the conserved Gly154 of subtilisin E: isolation and characterization of the revertant ...
|
|
52124
|
Random mutagenesis into the conserved Gly154 of subtilisin E: isolation and characterization of the revertant ...
|
|
52125
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52126
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52127
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52128
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52129
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52130
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52131
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52132
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52133
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52134
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52135
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52136
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52137
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52138
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52139
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52140
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52141
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52142
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52143
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52144
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52145
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52146
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52147
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52148
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52149
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52150
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52151
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52152
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52153
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52154
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52155
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52156
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52157
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52158
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52159
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52160
|
Properties of recombinant human cytosolic sialidase HsNEU2. The enzyme hydrolyzes monomerically dispersed GM1 ...
|
|
52161
|
The Bacillus subtilis counterpart of the mammalian 3-methyladenine DNA glycosylase has hypoxanthine and ...
|
|
52162
|
The Bacillus subtilis counterpart of the mammalian 3-methyladenine DNA glycosylase has hypoxanthine and ...
|
|
52163
|
The Bacillus subtilis counterpart of the mammalian 3-methyladenine DNA glycosylase has hypoxanthine and ...
|
|
52164
|
The Bacillus subtilis counterpart of the mammalian 3-methyladenine DNA glycosylase has hypoxanthine and ...
|
|
52165
|
Site-directed mutagenesis of human membrane-associated ganglioside sialidase: identification of amino-acid ...
|
|
52166
|
Site-directed mutagenesis of human membrane-associated ganglioside sialidase: identification of amino-acid ...
|
|
52167
|
Site-directed mutagenesis of human membrane-associated ganglioside sialidase: identification of amino-acid ...
|
|
52168
|
Site-directed mutagenesis of human membrane-associated ganglioside sialidase: identification of amino-acid ...
|
|
52169
|
Site-directed mutagenesis of human membrane-associated ganglioside sialidase: identification of amino-acid ...
|
|
52170
|
Site-directed mutagenesis of human membrane-associated ganglioside sialidase: identification of amino-acid ...
|
|
52171
|
Site-directed mutagenesis of human membrane-associated ganglioside sialidase: identification of amino-acid ...
|
|
52172
|
Site-directed mutagenesis of human membrane-associated ganglioside sialidase: identification of amino-acid ...
|
|
52173
|
Site-directed mutagenesis of human membrane-associated ganglioside sialidase: identification of amino-acid ...
|
|
52174
|
Site-directed mutagenesis of human membrane-associated ganglioside sialidase: identification of amino-acid ...
|
|
52175
|
Site-directed mutagenesis of human membrane-associated ganglioside sialidase: identification of amino-acid ...
|
|
52176
|
Site-directed mutagenesis of human membrane-associated ganglioside sialidase: identification of amino-acid ...
|
|
52177
|
Site-directed mutagenesis of human membrane-associated ganglioside sialidase: identification of amino-acid ...
|
|
52178
|
Site-directed mutagenesis of human membrane-associated ganglioside sialidase: identification of amino-acid ...
|
|
52179
|
Site-directed mutagenesis of human membrane-associated ganglioside sialidase: identification of amino-acid ...
|
|
52180
|
Site-directed mutagenesis of human membrane-associated ganglioside sialidase: identification of amino-acid ...
|
|
52181
|
Site-directed mutagenesis of human membrane-associated ganglioside sialidase: identification of amino-acid ...
|
|
52182
|
Inhibition of cholesterol biosynthesis in 3T3 fibroblasts by 2-aza-2,3-dihydrosqualene, a rationally designed ...
|
|
52183
|
Inhibition of cholesterol biosynthesis in 3T3 fibroblasts by 2-aza-2,3-dihydrosqualene, a rationally designed ...
|
|
52184
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52185
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52186
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52187
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52188
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52189
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52190
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52191
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52192
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52193
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52194
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52195
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52196
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52197
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52198
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52199
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52200
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52201
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52202
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52203
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52204
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52205
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52206
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52207
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52208
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52209
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52210
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52211
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52212
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52213
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52214
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52215
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52216
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52217
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52218
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52219
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52220
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52221
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52222
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52223
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52224
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52225
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52226
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52227
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52228
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52229
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52230
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52231
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52232
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52233
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52234
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52235
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52236
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52237
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52238
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52239
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52240
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52241
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52242
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52243
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52244
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52245
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52246
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52247
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52248
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52249
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52250
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52251
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52252
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52253
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52254
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52255
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52256
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52257
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52258
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52259
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52260
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52261
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52262
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52263
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52264
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52265
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52266
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52267
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52268
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52269
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52270
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52271
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52272
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52273
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52274
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52275
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52276
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52277
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52278
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52279
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52280
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52281
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52282
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52283
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52284
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52285
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52286
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52287
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52288
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52289
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52290
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52291
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52292
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52293
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52294
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52295
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52296
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52297
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52298
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52299
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52300
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52301
|
Structure-based design and synthesis of lipophilic 2,4-diamino-6-substituted quinazolines and their evaluation ...
|
|
52302
|
The recognition of glycolate oxidase apoprotein with flavin analogs in higher plants.
|
|
52303
|
The recognition of glycolate oxidase apoprotein with flavin analogs in higher plants.
|
|
52304
|
Characterization of Ke 6, a new 17beta-hydroxysteroid dehydrogenase, and its expression in gonadal tissues
|
|
52305
|
Characterization of Ke 6, a new 17beta-hydroxysteroid dehydrogenase, and its expression in gonadal tissues
|
|
52306
|
Characterization of Ke 6, a new 17beta-hydroxysteroid dehydrogenase, and its expression in gonadal tissues
|
|
52307
|
Characterization of Ke 6, a new 17beta-hydroxysteroid dehydrogenase, and its expression in gonadal tissues
|
|
52308
|
The functional glycosyltransferase signature sequence of the human beta 1,3-glucuronosyltransferase is a XDD ...
|
|
52309
|
The functional glycosyltransferase signature sequence of the human beta 1,3-glucuronosyltransferase is a XDD ...
|
|
52310
|
The functional glycosyltransferase signature sequence of the human beta 1,3-glucuronosyltransferase is a XDD ...
|
|
52311
|
Single amino acid substitutions disrupt tetramer formation in the dihydroneopterin aldolase enzyme of ...
|
|
52312
|
Substrate specificity of aminopeptidase Ey from hen's (Gallus domesticus) egg yolk
|
|
52313
|
The formation of uridine diphosphate-glucuronic acid in plants. Uridine diphosphate-glucuronic acid ...
|
|
52314
|
The formation of uridine diphosphate-glucuronic acid in plants. Uridine diphosphate-glucuronic acid ...
|
|
52315
|
Glutamate synthase: identification of the NADPH-binding site by site-directed mutagenesis
|
|
52316
|
Glutamate synthase: identification of the NADPH-binding site by site-directed mutagenesis
|
|
52317
|
Glutamate synthase: identification of the NADPH-binding site by site-directed mutagenesis
|
|
52318
|
Glutamate synthase: identification of the NADPH-binding site by site-directed mutagenesis
|
|
52319
|
Glutamate synthase: identification of the NADPH-binding site by site-directed mutagenesis
|
|
52320
|
Glutamate synthase: identification of the NADPH-binding site by site-directed mutagenesis
|
|
52321
|
Glutamate synthase: identification of the NADPH-binding site by site-directed mutagenesis
|
|
52322
|
Glutamate synthase: identification of the NADPH-binding site by site-directed mutagenesis
|
|
52323
|
Activity and stability of native and modified subtilisins in various media
|
|
52324
|
Activity and stability of native and modified subtilisins in various media
|
|
52325
|
Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity
|
|
52326
|
Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity
|
|
52327
|
Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity
|
|
52328
|
Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity
|
|
52329
|
The purification and properties of sucrose synthetase from etiolated Phaseolus aureus seedlings.
|
|
52330
|
The purification and properties of sucrose synthetase from etiolated Phaseolus aureus seedlings.
|
|
52331
|
The purification and properties of sucrose synthetase from etiolated Phaseolus aureus seedlings.
|
|
52332
|
The purification and properties of sucrose synthetase from etiolated Phaseolus aureus seedlings.
|
|
52333
|
The purification and properties of sucrose synthetase from etiolated Phaseolus aureus seedlings.
|
|
52334
|
The purification and properties of sucrose synthetase from etiolated Phaseolus aureus seedlings.
|
|
52335
|
The purification and properties of sucrose synthetase from etiolated Phaseolus aureus seedlings.
|
|
52336
|
The purification and properties of sucrose synthetase from etiolated Phaseolus aureus seedlings.
|
|
52337
|
A strategically positioned cation is crucial for efficient catalysis by chorismate mutase
|
|
52338
|
A strategically positioned cation is crucial for efficient catalysis by chorismate mutase
|
|
52339
|
A strategically positioned cation is crucial for efficient catalysis by chorismate mutase
|
|
52340
|
A strategically positioned cation is crucial for efficient catalysis by chorismate mutase
|
|
52341
|
A strategically positioned cation is crucial for efficient catalysis by chorismate mutase
|
|
52342
|
A strategically positioned cation is crucial for efficient catalysis by chorismate mutase
|
|
52343
|
Studies on the use of sepharose-N-(6-aminohexanoyl)-2-amino-2-deoxy-D-glucopyranose for the large-scale ...
|
|
52344
|
Studies on the use of sepharose-N-(6-aminohexanoyl)-2-amino-2-deoxy-D-glucopyranose for the large-scale ...
|
|
52345
|
Studies on the use of sepharose-N-(6-aminohexanoyl)-2-amino-2-deoxy-D-glucopyranose for the large-scale ...
|
|
52346
|
Identification and characterization of a plastid-localized Arabidopsis glyoxylate reductase isoform: ...
|
|
52347
|
Identification and characterization of a plastid-localized Arabidopsis glyoxylate reductase isoform: ...
|
|
52348
|
Identification and characterization of a plastid-localized Arabidopsis glyoxylate reductase isoform: ...
|
|
52349
|
Identification and characterization of a plastid-localized Arabidopsis glyoxylate reductase isoform: ...
|
|
52350
|
Cloning of Glucuronokinase from Arabidopsis thaliana, the last missing enzyme of the myo-inositol oxygenase ...
|
|
52351
|
Cloning of Glucuronokinase from Arabidopsis thaliana, the last missing enzyme of the myo-inositol oxygenase ...
|
|
52352
|
The importance of the strictly conserved, C-terminal glycine residue in phosphoenolpyruvate carboxylase for ...
|
|
52353
|
The importance of the strictly conserved, C-terminal glycine residue in phosphoenolpyruvate carboxylase for ...
|
|
52354
|
The importance of the strictly conserved, C-terminal glycine residue in phosphoenolpyruvate carboxylase for ...
|
|
52355
|
The importance of the strictly conserved, C-terminal glycine residue in phosphoenolpyruvate carboxylase for ...
|
|
52356
|
The importance of the strictly conserved, C-terminal glycine residue in phosphoenolpyruvate carboxylase for ...
|
|
52357
|
A highly specific L-galactose-1-phosphate phosphatase on the path to ascorbate biosynthesis
|
|
52358
|
A highly specific L-galactose-1-phosphate phosphatase on the path to ascorbate biosynthesis
|
|
52359
|
A highly specific L-galactose-1-phosphate phosphatase on the path to ascorbate biosynthesis
|
|
52360
|
A highly specific L-galactose-1-phosphate phosphatase on the path to ascorbate biosynthesis
|
|
52361
|
A highly specific L-galactose-1-phosphate phosphatase on the path to ascorbate biosynthesis
|
|
52362
|
A highly specific L-galactose-1-phosphate phosphatase on the path to ascorbate biosynthesis
|
|
52363
|
A highly specific L-galactose-1-phosphate phosphatase on the path to ascorbate biosynthesis
|
|
52364
|
A highly specific L-galactose-1-phosphate phosphatase on the path to ascorbate biosynthesis
|
|
52365
|
A highly specific L-galactose-1-phosphate phosphatase on the path to ascorbate biosynthesis
|
|
52366
|
A highly specific L-galactose-1-phosphate phosphatase on the path to ascorbate biosynthesis
|
|
52367
|
A highly specific L-galactose-1-phosphate phosphatase on the path to ascorbate biosynthesis
|
|
52368
|
A highly specific L-galactose-1-phosphate phosphatase on the path to ascorbate biosynthesis
|
|
52369
|
Probing the role of cysteine residues in glucosamine-1-phosphate acetyltransferase activity of the ...
|
|
52370
|
Probing the role of cysteine residues in glucosamine-1-phosphate acetyltransferase activity of the ...
|
|
52371
|
Probing the role of cysteine residues in glucosamine-1-phosphate acetyltransferase activity of the ...
|
|
52372
|
Probing the role of cysteine residues in glucosamine-1-phosphate acetyltransferase activity of the ...
|
|
52373
|
Probing the role of cysteine residues in glucosamine-1-phosphate acetyltransferase activity of the ...
|
|
52374
|
Probing the role of cysteine residues in glucosamine-1-phosphate acetyltransferase activity of the ...
|
|
52375
|
Probing the role of cysteine residues in glucosamine-1-phosphate acetyltransferase activity of the ...
|
|
52376
|
Probing the role of cysteine residues in glucosamine-1-phosphate acetyltransferase activity of the ...
|
|
52377
|
Probing the role of cysteine residues in glucosamine-1-phosphate acetyltransferase activity of the ...
|
|
52378
|
Probing the role of cysteine residues in glucosamine-1-phosphate acetyltransferase activity of the ...
|
|
52379
|
Probing the role of cysteine residues in glucosamine-1-phosphate acetyltransferase activity of the ...
|
|
52380
|
Probing the role of cysteine residues in glucosamine-1-phosphate acetyltransferase activity of the ...
|
|
52381
|
Probing the role of cysteine residues in glucosamine-1-phosphate acetyltransferase activity of the ...
|
|
52382
|
Probing the role of cysteine residues in glucosamine-1-phosphate acetyltransferase activity of the ...
|
|
52383
|
A novel polymorphic allele of human arylacetamide deacetylase leads to decreased enzyme activity
|
|
52384
|
A novel polymorphic allele of human arylacetamide deacetylase leads to decreased enzyme activity
|
|
52385
|
A novel polymorphic allele of human arylacetamide deacetylase leads to decreased enzyme activity
|
|
52386
|
A novel polymorphic allele of human arylacetamide deacetylase leads to decreased enzyme activity
|
|
52387
|
A novel polymorphic allele of human arylacetamide deacetylase leads to decreased enzyme activity
|
|
52388
|
A novel polymorphic allele of human arylacetamide deacetylase leads to decreased enzyme activity
|
|
52389
|
A novel polymorphic allele of human arylacetamide deacetylase leads to decreased enzyme activity
|
|
52390
|
A novel polymorphic allele of human arylacetamide deacetylase leads to decreased enzyme activity
|
|
52391
|
A novel polymorphic allele of human arylacetamide deacetylase leads to decreased enzyme activity
|
|
52392
|
Cloning and characterization of the glucooligosaccharide catabolic pathway beta-glucan glucohydrolase and ...
|
|
52393
|
Cloning and characterization of the glucooligosaccharide catabolic pathway beta-glucan glucohydrolase and ...
|
|
52394
|
Cloning and characterization of the glucooligosaccharide catabolic pathway beta-glucan glucohydrolase and ...
|
|
52395
|
Cloning and characterization of the glucooligosaccharide catabolic pathway beta-glucan glucohydrolase and ...
|
|
52396
|
Cloning and characterization of the glucooligosaccharide catabolic pathway beta-glucan glucohydrolase and ...
|
|
52397
|
Cloning and characterization of the glucooligosaccharide catabolic pathway beta-glucan glucohydrolase and ...
|
|
52398
|
Cool C4 photosynthesis: pyruvate Pi dikinase expression and activity corresponds to the exceptional cold ...
|
|
52399
|
Cool C4 photosynthesis: pyruvate Pi dikinase expression and activity corresponds to the exceptional cold ...
|
|
52400
|
Cool C4 photosynthesis: pyruvate Pi dikinase expression and activity corresponds to the exceptional cold ...
|
|
52401
|
Cool C4 photosynthesis: pyruvate Pi dikinase expression and activity corresponds to the exceptional cold ...
|
|
52402
|
Cool C4 photosynthesis: pyruvate Pi dikinase expression and activity corresponds to the exceptional cold ...
|
|
52403
|
Cool C4 photosynthesis: pyruvate Pi dikinase expression and activity corresponds to the exceptional cold ...
|
|
52404
|
Cool C4 photosynthesis: pyruvate Pi dikinase expression and activity corresponds to the exceptional cold ...
|
|
52405
|
Cool C4 photosynthesis: pyruvate Pi dikinase expression and activity corresponds to the exceptional cold ...
|
|
52406
|
Cool C4 photosynthesis: pyruvate Pi dikinase expression and activity corresponds to the exceptional cold ...
|
|
52407
|
Cool C4 photosynthesis: pyruvate Pi dikinase expression and activity corresponds to the exceptional cold ...
|
|
52408
|
Cool C4 photosynthesis: pyruvate Pi dikinase expression and activity corresponds to the exceptional cold ...
|
|
52409
|
Cool C4 photosynthesis: pyruvate Pi dikinase expression and activity corresponds to the exceptional cold ...
|
|
52410
|
Cool C4 photosynthesis: pyruvate Pi dikinase expression and activity corresponds to the exceptional cold ...
|
|
52411
|
Cool C4 photosynthesis: pyruvate Pi dikinase expression and activity corresponds to the exceptional cold ...
|
|
52412
|
Cool C4 photosynthesis: pyruvate Pi dikinase expression and activity corresponds to the exceptional cold ...
|
|
52413
|
Cool C4 photosynthesis: pyruvate Pi dikinase expression and activity corresponds to the exceptional cold ...
|
|
52414
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52415
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52416
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52417
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52418
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52419
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52420
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52421
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52422
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52423
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52424
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52425
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52426
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52427
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52428
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52429
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52430
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52431
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52432
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52433
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52434
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52435
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52436
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52437
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52438
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52439
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52440
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52441
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52442
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52443
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52444
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52445
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52446
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52447
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52448
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52449
|
The heterogeneity of mast cell tryptase from human lung and skin
|
|
52450
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52451
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52452
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52453
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52454
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52455
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52456
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52457
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52458
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52459
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52460
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52461
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52462
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52463
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52464
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52465
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52466
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52467
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52468
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52469
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52470
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52471
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52472
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52473
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52474
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52475
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52476
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52477
|
Domain-specific determinants of catalysis/substrate binding and the oligomerization status of barley ...
|
|
52478
|
Sucrose metabolism in cyanobacteria: sucrose synthase from Anabaena sp. strain PCC 7119 is remarkably ...
|
|
52479
|
Sucrose metabolism in cyanobacteria: sucrose synthase from Anabaena sp. strain PCC 7119 is remarkably ...
|
|
52480
|
Sucrose metabolism in cyanobacteria: sucrose synthase from Anabaena sp. strain PCC 7119 is remarkably ...
|
|
52481
|
Sucrose metabolism in cyanobacteria: sucrose synthase from Anabaena sp. strain PCC 7119 is remarkably ...
|
|
52482
|
Sucrose metabolism in cyanobacteria: sucrose synthase from Anabaena sp. strain PCC 7119 is remarkably ...
|
|
52483
|
Sucrose metabolism in cyanobacteria: sucrose synthase from Anabaena sp. strain PCC 7119 is remarkably ...
|
|
52484
|
Sucrose metabolism in cyanobacteria: sucrose synthase from Anabaena sp. strain PCC 7119 is remarkably ...
|
|
52485
|
Sucrose metabolism in cyanobacteria: sucrose synthase from Anabaena sp. strain PCC 7119 is remarkably ...
|
|
52486
|
Sucrose metabolism in cyanobacteria: sucrose synthase from Anabaena sp. strain PCC 7119 is remarkably ...
|
|
52487
|
Sucrose metabolism in cyanobacteria: sucrose synthase from Anabaena sp. strain PCC 7119 is remarkably ...
|
|
52488
|
Sucrose metabolism in cyanobacteria: sucrose synthase from Anabaena sp. strain PCC 7119 is remarkably ...
|
|
52489
|
Sucrose metabolism in cyanobacteria: sucrose synthase from Anabaena sp. strain PCC 7119 is remarkably ...
|
|
52490
|
Sucrose metabolism in cyanobacteria: sucrose synthase from Anabaena sp. strain PCC 7119 is remarkably ...
|
|
52491
|
Sucrose metabolism in cyanobacteria: sucrose synthase from Anabaena sp. strain PCC 7119 is remarkably ...
|
|
52492
|
Sucrose metabolism in cyanobacteria: sucrose synthase from Anabaena sp. strain PCC 7119 is remarkably ...
|
|
52493
|
Sucrose metabolism in cyanobacteria: sucrose synthase from Anabaena sp. strain PCC 7119 is remarkably ...
|
|
52494
|
Sucrose metabolism in cyanobacteria: sucrose synthase from Anabaena sp. strain PCC 7119 is remarkably ...
|
|
52495
|
Sucrose metabolism in cyanobacteria: sucrose synthase from Anabaena sp. strain PCC 7119 is remarkably ...
|
|
52496
|
Sucrose metabolism in cyanobacteria: sucrose synthase from Anabaena sp. strain PCC 7119 is remarkably ...
|
|
52497
|
Sucrose metabolism in cyanobacteria: sucrose synthase from Anabaena sp. strain PCC 7119 is remarkably ...
|
|
52498
|
Comparative analyses of hairpin substrate recognition by Escherichia coli and Bacillus subtilis ribonuclease P ...
|
|
52499
|
Comparative analyses of hairpin substrate recognition by Escherichia coli and Bacillus subtilis ribonuclease P ...
|
|
52500
|
Comparative analyses of hairpin substrate recognition by Escherichia coli and Bacillus subtilis ribonuclease P ...
|
|
52501
|
Comparative analyses of hairpin substrate recognition by Escherichia coli and Bacillus subtilis ribonuclease P ...
|
|
52502
|
Comparative analyses of hairpin substrate recognition by Escherichia coli and Bacillus subtilis ribonuclease P ...
|
|
52503
|
Comparative analyses of hairpin substrate recognition by Escherichia coli and Bacillus subtilis ribonuclease P ...
|
|
52504
|
Post-translational processing, metabolic stability and catalytic efficiency of oat arginine decarboxylase ...
|
|
52505
|
Post-translational processing, metabolic stability and catalytic efficiency of oat arginine decarboxylase ...
|
|
52506
|
Arabidopsis thaliana NADP-malic enzyme isoforms: high degree of identity but clearly distinct properties
|
|
52507
|
Arabidopsis thaliana NADP-malic enzyme isoforms: high degree of identity but clearly distinct properties
|
|
52508
|
Arabidopsis thaliana NADP-malic enzyme isoforms: high degree of identity but clearly distinct properties
|
|
52509
|
Arabidopsis thaliana NADP-malic enzyme isoforms: high degree of identity but clearly distinct properties
|
|
52510
|
Arabidopsis thaliana NADP-malic enzyme isoforms: high degree of identity but clearly distinct properties
|
|
52511
|
Arabidopsis thaliana NADP-malic enzyme isoforms: high degree of identity but clearly distinct properties
|
|
52512
|
Arabidopsis thaliana NADP-malic enzyme isoforms: high degree of identity but clearly distinct properties
|
|
52513
|
Arabidopsis thaliana NADP-malic enzyme isoforms: high degree of identity but clearly distinct properties
|
|
52514
|
Arabidopsis thaliana NADP-malic enzyme isoforms: high degree of identity but clearly distinct properties
|
|
52515
|
Arabidopsis thaliana NADP-malic enzyme isoforms: high degree of identity but clearly distinct properties
|
|
52516
|
Arabidopsis thaliana NADP-malic enzyme isoforms: high degree of identity but clearly distinct properties
|
|
52517
|
Arabidopsis thaliana NADP-malic enzyme isoforms: high degree of identity but clearly distinct properties
|
|
52518
|
Arabidopsis thaliana NADP-malic enzyme isoforms: high degree of identity but clearly distinct properties
|
|
52519
|
Arabidopsis thaliana NADP-malic enzyme isoforms: high degree of identity but clearly distinct properties
|
|
52520
|
Arabidopsis thaliana NADP-malic enzyme isoforms: high degree of identity but clearly distinct properties
|
|
52521
|
Arabidopsis thaliana NADP-malic enzyme isoforms: high degree of identity but clearly distinct properties
|
|
52522
|
Arabidopsis thaliana NADP-malic enzyme isoforms: high degree of identity but clearly distinct properties
|
|
52523
|
Arabidopsis thaliana NADP-malic enzyme isoforms: high degree of identity but clearly distinct properties
|
|
52524
|
Arabidopsis thaliana NADP-malic enzyme isoforms: high degree of identity but clearly distinct properties
|
|
52525
|
Arabidopsis thaliana NADP-malic enzyme isoforms: high degree of identity but clearly distinct properties
|
|
52526
|
Arabidopsis thaliana NADP-malic enzyme isoforms: high degree of identity but clearly distinct properties
|
|
52527
|
Arabidopsis thaliana NADP-malic enzyme isoforms: high degree of identity but clearly distinct properties
|
|
52528
|
Hydrolysis of N-succinyl-L,L-diaminopimelic acid by the Haemophilus influenzae dapE-encoded desuccinylase: ...
|
|
52529
|
Hydrolysis of N-succinyl-L,L-diaminopimelic acid by the Haemophilus influenzae dapE-encoded desuccinylase: ...
|
|
52530
|
Hydrolysis of N-succinyl-L,L-diaminopimelic acid by the Haemophilus influenzae dapE-encoded desuccinylase: ...
|
|
52531
|
Hydrolysis of N-succinyl-L,L-diaminopimelic acid by the Haemophilus influenzae dapE-encoded desuccinylase: ...
|
|
52532
|
Hydrolysis of N-succinyl-L,L-diaminopimelic acid by the Haemophilus influenzae dapE-encoded desuccinylase: ...
|
|
52533
|
Hydrolysis of N-succinyl-L,L-diaminopimelic acid by the Haemophilus influenzae dapE-encoded desuccinylase: ...
|
|
52534
|
Hydrolysis of N-succinyl-L,L-diaminopimelic acid by the Haemophilus influenzae dapE-encoded desuccinylase: ...
|
|
52535
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52536
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52537
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52538
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52539
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52540
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52541
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52542
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52543
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52544
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52545
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52546
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52547
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52548
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52549
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52550
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52551
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52552
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52553
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52554
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52555
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52556
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52557
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52558
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52559
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52560
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52561
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52562
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52563
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52564
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52565
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52566
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52567
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52568
|
An O-glycoside of sialic acid derivative that inhibits both hemagglutinin and sialidase activities of ...
|
|
52569
|
Identification, characterization and functional expression of a tyrosine ammonia-lyase and its mutants from ...
|
|
52570
|
Identification, characterization and functional expression of a tyrosine ammonia-lyase and its mutants from ...
|
|
52571
|
Identification, characterization and functional expression of a tyrosine ammonia-lyase and its mutants from ...
|
|
52572
|
Identification, characterization and functional expression of a tyrosine ammonia-lyase and its mutants from ...
|
|
52573
|
Identification, characterization and functional expression of a tyrosine ammonia-lyase and its mutants from ...
|
|
52574
|
New class of bacterial phenylalanyl-tRNA synthetase inhibitors with high potency and broad-spectrum activity
|
|
52575
|
New class of bacterial phenylalanyl-tRNA synthetase inhibitors with high potency and broad-spectrum activity
|
|
52576
|
Characterization of two phosphate transporters from barley; evidence for diverse function and kinetic ...
|
|
52577
|
Characterization of two phosphate transporters from barley; evidence for diverse function and kinetic ...
|
|
52578
|
Characterization of two phosphate transporters from barley; evidence for diverse function and kinetic ...
|
|
52579
|
Characterization of two phosphate transporters from barley; evidence for diverse function and kinetic ...
|
|
52580
|
Why boys will be boys: two pathways of fetal testicular androgen biosynthesis are needed for male sexual ...
|
|
52581
|
Why boys will be boys: two pathways of fetal testicular androgen biosynthesis are needed for male sexual ...
|
|
52582
|
Why boys will be boys: two pathways of fetal testicular androgen biosynthesis are needed for male sexual ...
|
|
52583
|
Why boys will be boys: two pathways of fetal testicular androgen biosynthesis are needed for male sexual ...
|
|
52584
|
Why boys will be boys: two pathways of fetal testicular androgen biosynthesis are needed for male sexual ...
|
|
52585
|
Why boys will be boys: two pathways of fetal testicular androgen biosynthesis are needed for male sexual ...
|
|
52586
|
Why boys will be boys: two pathways of fetal testicular androgen biosynthesis are needed for male sexual ...
|
|
52587
|
Why boys will be boys: two pathways of fetal testicular androgen biosynthesis are needed for male sexual ...
|
|
52588
|
Why boys will be boys: two pathways of fetal testicular androgen biosynthesis are needed for male sexual ...
|
|
52589
|
Why boys will be boys: two pathways of fetal testicular androgen biosynthesis are needed for male sexual ...
|
|
52590
|
Why boys will be boys: two pathways of fetal testicular androgen biosynthesis are needed for male sexual ...
|
|
52591
|
Identification and functional analysis of a prokaryotic-type aspartate aminotransferase: implications for ...
|
|
52592
|
Identification and functional analysis of a prokaryotic-type aspartate aminotransferase: implications for ...
|
|
52593
|
Crystal structure of human MTH1 and the 8-oxo-dGMP product complex.
|
|
52594
|
The oxidized forms of dATP are substrates for the human MutT homologue, the hMTH1 protein
|
|
52595
|
The oxidized forms of dATP are substrates for the human MutT homologue, the hMTH1 protein
|
|
52596
|
The oxidized forms of dATP are substrates for the human MutT homologue, the hMTH1 protein
|
|
52597
|
The oxidized forms of dATP are substrates for the human MutT homologue, the hMTH1 protein
|
|
52598
|
The oxidized forms of dATP are substrates for the human MutT homologue, the hMTH1 protein
|
|
52599
|
The oxidized forms of dATP are substrates for the human MutT homologue, the hMTH1 protein
|
|
52600
|
The oxidized forms of dATP are substrates for the human MutT homologue, the hMTH1 protein
|
|
52601
|
The oxidized forms of dATP are substrates for the human MutT homologue, the hMTH1 protein
|
|
52602
|
The oxidized forms of dATP are substrates for the human MutT homologue, the hMTH1 protein
|
|
52603
|
The oxidized forms of dATP are substrates for the human MutT homologue, the hMTH1 protein
|
|
52604
|
The oxidized forms of dATP are substrates for the human MutT homologue, the hMTH1 protein
|
|
52605
|
The oxidized forms of dATP are substrates for the human MutT homologue, the hMTH1 protein
|
|
52606
|
Inhibitors of phenylalanine ammonia-lyase: substituted derivatives of 2-aminoindane-2-phosphonic acid and ...
|
|
52607
|
Inhibitors of phenylalanine ammonia-lyase: substituted derivatives of 2-aminoindane-2-phosphonic acid and ...
|
|
52608
|
Inhibitors of phenylalanine ammonia-lyase: substituted derivatives of 2-aminoindane-2-phosphonic acid and ...
|
|
52609
|
Inhibitors of phenylalanine ammonia-lyase: substituted derivatives of 2-aminoindane-2-phosphonic acid and ...
|
|
52610
|
Kinetic analysis of the reactions catalyzed by histidine and phenylalanine ammonia lyases
|
|
52611
|
Kinetic analysis of the reactions catalyzed by histidine and phenylalanine ammonia lyases
|
|
52612
|
Kinetic analysis of the reactions catalyzed by histidine and phenylalanine ammonia lyases
|
|
52613
|
Kinetic analysis of the reactions catalyzed by histidine and phenylalanine ammonia lyases
|
|
52614
|
Kinetic analysis of the reactions catalyzed by histidine and phenylalanine ammonia lyases
|
|
52615
|
Kinetic analysis of the reactions catalyzed by histidine and phenylalanine ammonia lyases
|
|
52616
|
Kinetic analysis of the reactions catalyzed by histidine and phenylalanine ammonia lyases
|
|
52617
|
Kinetic analysis of the reactions catalyzed by histidine and phenylalanine ammonia lyases
|
|
52618
|
Examination of the role of Gln-158 in the mechanism of CO(2) hydration catalyzed by beta-carbonic anhydrase ...
|
|
52619
|
Bacillus subtilis isocitrate dehydrogenase. A substrate analogue for Escherichia coli isocitrate dehydrogenase ...
|
|
52620
|
Bacillus subtilis isocitrate dehydrogenase. A substrate analogue for Escherichia coli isocitrate dehydrogenase ...
|
|
52621
|
Bacillus subtilis isocitrate dehydrogenase. A substrate analogue for Escherichia coli isocitrate dehydrogenase ...
|
|
52622
|
Bacillus subtilis isocitrate dehydrogenase. A substrate analogue for Escherichia coli isocitrate dehydrogenase ...
|
|
52623
|
Bacillus subtilis isocitrate dehydrogenase. A substrate analogue for Escherichia coli isocitrate dehydrogenase ...
|
|
52624
|
Bacillus subtilis isocitrate dehydrogenase. A substrate analogue for Escherichia coli isocitrate dehydrogenase ...
|
|
52625
|
Distinct properties of the five UDP-D-glucose/UDP-D-galactose 4-epimerase isoforms of Arabidopsis thaliana
|
|
52626
|
Distinct properties of the five UDP-D-glucose/UDP-D-galactose 4-epimerase isoforms of Arabidopsis thaliana
|
|
52627
|
Distinct properties of the five UDP-D-glucose/UDP-D-galactose 4-epimerase isoforms of Arabidopsis thaliana
|
|
52628
|
Distinct properties of the five UDP-D-glucose/UDP-D-galactose 4-epimerase isoforms of Arabidopsis thaliana
|
|
52629
|
Distinct properties of the five UDP-D-glucose/UDP-D-galactose 4-epimerase isoforms of Arabidopsis thaliana
|
|
52630
|
Distinct properties of the five UDP-D-glucose/UDP-D-galactose 4-epimerase isoforms of Arabidopsis thaliana
|
|
52631
|
Distinct properties of the five UDP-D-glucose/UDP-D-galactose 4-epimerase isoforms of Arabidopsis thaliana
|
|
52632
|
Distinct properties of the five UDP-D-glucose/UDP-D-galactose 4-epimerase isoforms of Arabidopsis thaliana
|
|
52633
|
Distinct properties of the five UDP-D-glucose/UDP-D-galactose 4-epimerase isoforms of Arabidopsis thaliana
|
|
52634
|
Distinct properties of the five UDP-D-glucose/UDP-D-galactose 4-epimerase isoforms of Arabidopsis thaliana
|
|
52635
|
Enzymic mechanism of starch stynthesis in ripening rice grains. VII. Purification and enzymic properties of ...
|
|
52636
|
Enzymic mechanism of starch stynthesis in ripening rice grains. VII. Purification and enzymic properties of ...
|
|
52637
|
Enzymic mechanism of starch stynthesis in ripening rice grains. VII. Purification and enzymic properties of ...
|
|
52638
|
Enzymic mechanism of starch stynthesis in ripening rice grains. VII. Purification and enzymic properties of ...
|
|
52639
|
Enzymic mechanism of starch stynthesis in ripening rice grains. VII. Purification and enzymic properties of ...
|
|
52640
|
Enzymic mechanism of starch stynthesis in ripening rice grains. VII. Purification and enzymic properties of ...
|
|
52641
|
Enzymic mechanism of starch stynthesis in ripening rice grains. VII. Purification and enzymic properties of ...
|
|
52642
|
Enzymic mechanism of starch stynthesis in ripening rice grains. VII. Purification and enzymic properties of ...
|
|
52643
|
Structure of Arabidopsis dehydroquinate dehydratase-shikimate dehydrogenase and implications for metabolic ...
|
|
52644
|
Structure of Arabidopsis dehydroquinate dehydratase-shikimate dehydrogenase and implications for metabolic ...
|
|
52645
|
Structure of Arabidopsis dehydroquinate dehydratase-shikimate dehydrogenase and implications for metabolic ...
|
|
52646
|
Structure of Arabidopsis dehydroquinate dehydratase-shikimate dehydrogenase and implications for metabolic ...
|
|
52647
|
Structure of Arabidopsis dehydroquinate dehydratase-shikimate dehydrogenase and implications for metabolic ...
|
|
52648
|
Structure of Arabidopsis dehydroquinate dehydratase-shikimate dehydrogenase and implications for metabolic ...
|
|
52649
|
Structure of Arabidopsis dehydroquinate dehydratase-shikimate dehydrogenase and implications for metabolic ...
|
|
52650
|
Structure of Arabidopsis dehydroquinate dehydratase-shikimate dehydrogenase and implications for metabolic ...
|
|
52651
|
Structure of Arabidopsis dehydroquinate dehydratase-shikimate dehydrogenase and implications for metabolic ...
|
|
52652
|
Structure of Arabidopsis dehydroquinate dehydratase-shikimate dehydrogenase and implications for metabolic ...
|
|
52653
|
Structure of Arabidopsis dehydroquinate dehydratase-shikimate dehydrogenase and implications for metabolic ...
|
|
52654
|
Structure of Arabidopsis dehydroquinate dehydratase-shikimate dehydrogenase and implications for metabolic ...
|
|
52655
|
Structure of Arabidopsis dehydroquinate dehydratase-shikimate dehydrogenase and implications for metabolic ...
|
|
52656
|
Structure of Arabidopsis dehydroquinate dehydratase-shikimate dehydrogenase and implications for metabolic ...
|
|
52657
|
Structure of Arabidopsis dehydroquinate dehydratase-shikimate dehydrogenase and implications for metabolic ...
|
|
52658
|
Structure of Arabidopsis dehydroquinate dehydratase-shikimate dehydrogenase and implications for metabolic ...
|
|
52659
|
Structure of Arabidopsis dehydroquinate dehydratase-shikimate dehydrogenase and implications for metabolic ...
|
|
52660
|
Structure of Arabidopsis dehydroquinate dehydratase-shikimate dehydrogenase and implications for metabolic ...
|
|
52661
|
Structure of Arabidopsis dehydroquinate dehydratase-shikimate dehydrogenase and implications for metabolic ...
|
|
52662
|
Overexpression, purification, and stereochemical studies of the recombinant (S)-adenosyl-L-methionine: delta ...
|
|
52663
|
Overexpression, purification, and stereochemical studies of the recombinant (S)-adenosyl-L-methionine: delta ...
|
|
52664
|
Purification of and kinetic studies on a cloned protoporphyrinogen oxidase from the aerobic bacterium Bacillus ...
|
|
52665
|
Purification of and kinetic studies on a cloned protoporphyrinogen oxidase from the aerobic bacterium Bacillus ...
|
|
52666
|
Purification of and kinetic studies on a cloned protoporphyrinogen oxidase from the aerobic bacterium Bacillus ...
|
|
52667
|
Purification of and kinetic studies on a cloned protoporphyrinogen oxidase from the aerobic bacterium Bacillus ...
|
|
52668
|
Purification of and kinetic studies on a cloned protoporphyrinogen oxidase from the aerobic bacterium Bacillus ...
|
|
52669
|
Purification of and kinetic studies on a cloned protoporphyrinogen oxidase from the aerobic bacterium Bacillus ...
|
|
52670
|
Purification of and kinetic studies on a cloned protoporphyrinogen oxidase from the aerobic bacterium Bacillus ...
|
|
52671
|
Purification of and kinetic studies on a cloned protoporphyrinogen oxidase from the aerobic bacterium Bacillus ...
|
|
52672
|
Purification of and kinetic studies on a cloned protoporphyrinogen oxidase from the aerobic bacterium Bacillus ...
|
|
52673
|
Kinetic data Rostock S.pyogenes
|
|
52674
|
Kinetic data Rostock S.pyogenes
|
|
52675
|
Kinetic data Rostock S.pyogenes
|
|
52676
|
Kinetic data Rostock S.pyogenes
|
|
52677
|
Kinetic data Rostock S.pyogenes
|
|
52678
|
Kinetic data Rostock S.pyogenes
|
|
52679
|
Kinetic data Rostock S.pyogenes
|
|
52680
|
Kinetic data Rostock S.pyogenes
|
|
52681
|
Kinetic data Rostock E.faecalis
|
|
52682
|
Kinetic data Rostock E.faecalis
|
|
52683
|
Kinetic data Rostock E.faecalis
|
|
52684
|
Kinetic data Rostock E.faecalis
|
|
52685
|
Kinetic data Rostock E.faecalis
|
|
52686
|
Kinetic data Rostock E.faecalis
|
|
52687
|
Kinetic data Rostock E.faecalis
|
|
52688
|
Kinetic data Rostock E.faecalis
|
|
52689
|
Post-translational regulation of cytosolic glutamine synthetase of Medicago truncatula
|
|
52690
|
Post-translational regulation of cytosolic glutamine synthetase of Medicago truncatula
|
|
52691
|
Post-translational regulation of cytosolic glutamine synthetase of Medicago truncatula
|
|
52692
|
Post-translational regulation of cytosolic glutamine synthetase of Medicago truncatula
|
|
52693
|
Post-translational regulation of cytosolic glutamine synthetase of Medicago truncatula
|
|
52694
|
Post-translational regulation of cytosolic glutamine synthetase of Medicago truncatula
|
|
52695
|
Overproduction and characterization of the uracil-DNA glycosylase inhibitor of bacteriophage PBS2
|
|
52696
|
Overproduction and characterization of the uracil-DNA glycosylase inhibitor of bacteriophage PBS2
|
|
52697
|
Kinetic data Rostock L. plantarum
|
|
52698
|
Kinetic data Rostock L. plantarum
|
|
52699
|
Kinetic data Rostock L. plantarum
|
|
52700
|
Steady-state and transient kinetics of Escherichia coli nitric-oxide dioxygenase (flavohemoglobin). The B10 ...
|
|
52701
|
Steady-state and transient kinetics of Escherichia coli nitric-oxide dioxygenase (flavohemoglobin). The B10 ...
|
|
52702
|
Steady-state and transient kinetics of Escherichia coli nitric-oxide dioxygenase (flavohemoglobin). The B10 ...
|
|
52703
|
Steady-state and transient kinetics of Escherichia coli nitric-oxide dioxygenase (flavohemoglobin). The B10 ...
|
|
52704
|
Steady-state and transient kinetics of Escherichia coli nitric-oxide dioxygenase (flavohemoglobin). The B10 ...
|
|
52705
|
Steady-state and transient kinetics of Escherichia coli nitric-oxide dioxygenase (flavohemoglobin). The B10 ...
|
|
52706
|
Steady-state and transient kinetics of Escherichia coli nitric-oxide dioxygenase (flavohemoglobin). The B10 ...
|
|
52707
|
Steady-state and transient kinetics of Escherichia coli nitric-oxide dioxygenase (flavohemoglobin). The B10 ...
|
|
52708
|
Steady-state and transient kinetics of Escherichia coli nitric-oxide dioxygenase (flavohemoglobin). The B10 ...
|
|
52709
|
Steady-state and transient kinetics of Escherichia coli nitric-oxide dioxygenase (flavohemoglobin). The B10 ...
|
|
52710
|
Steady-state and transient kinetics of Escherichia coli nitric-oxide dioxygenase (flavohemoglobin). The B10 ...
|
|
52711
|
Steady-state and transient kinetics of Escherichia coli nitric-oxide dioxygenase (flavohemoglobin). The B10 ...
|
|
52712
|
Steady-state and transient kinetics of Escherichia coli nitric-oxide dioxygenase (flavohemoglobin). The B10 ...
|
|
52713
|
Steady-state and transient kinetics of Escherichia coli nitric-oxide dioxygenase (flavohemoglobin). The B10 ...
|
|
52714
|
Steady-state and transient kinetics of Escherichia coli nitric-oxide dioxygenase (flavohemoglobin). The B10 ...
|
|
52715
|
Steady-state and transient kinetics of Escherichia coli nitric-oxide dioxygenase (flavohemoglobin). The B10 ...
|
|
52716
|
Steady-state and transient kinetics of Escherichia coli nitric-oxide dioxygenase (flavohemoglobin). The B10 ...
|
|
52717
|
Steady-state and transient kinetics of Escherichia coli nitric-oxide dioxygenase (flavohemoglobin). The B10 ...
|
|
52718
|
Steady-state and transient kinetics of Escherichia coli nitric-oxide dioxygenase (flavohemoglobin). The B10 ...
|
|
52719
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52720
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52721
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52722
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52723
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52724
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52725
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52726
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52727
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52728
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52729
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52730
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52731
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52732
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52733
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52734
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52735
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52736
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52737
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52738
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52739
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52740
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52741
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52742
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52743
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52744
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52745
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52746
|
Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical ...
|
|
52747
|
ADP glucose pyrophosphorylase from maize endosperm.
|
|
52748
|
ADP glucose pyrophosphorylase from maize endosperm.
|
|
52749
|
ADP glucose pyrophosphorylase from maize endosperm.
|
|
52750
|
ADP glucose pyrophosphorylase from maize endosperm.
|
|
52751
|
ADP glucose pyrophosphorylase from maize endosperm.
|
|
52752
|
ADP glucose pyrophosphorylase from maize endosperm.
|
|
52753
|
ADP glucose pyrophosphorylase from maize endosperm.
|
|
52754
|
ADP glucose pyrophosphorylase from maize endosperm.
|
|
52755
|
ADP glucose pyrophosphorylase from maize endosperm.
|
|
52756
|
ADP glucose pyrophosphorylase from maize endosperm.
|
|
52757
|
ADP glucose pyrophosphorylase from maize endosperm.
|
|
52758
|
ADP glucose pyrophosphorylase from maize endosperm.
|
|
52759
|
ADP glucose pyrophosphorylase from maize endosperm.
|
|
52760
|
ADP glucose pyrophosphorylase from maize endosperm.
|
|
52761
|
ADP glucose pyrophosphorylase from maize endosperm.
|
|
52762
|
ADP glucose pyrophosphorylase from maize endosperm.
|
|
52763
|
ADP glucose pyrophosphorylase from maize endosperm.
|
|
52764
|
ADP glucose pyrophosphorylase from maize endosperm.
|
|
52765
|
ADP glucose pyrophosphorylase from maize endosperm.
|
|
52766
|
ADP glucose pyrophosphorylase from maize endosperm.
|
|
52767
|
ADP glucose pyrophosphorylase from maize endosperm.
|
|
52768
|
ADP glucose pyrophosphorylase from maize endosperm.
|
|
52769
|
ADP glucose pyrophosphorylase from maize endosperm.
|
|
52770
|
ADP glucose pyrophosphorylase from maize endosperm.
|
|
52771
|
ADP glucose pyrophosphorylase from maize endosperm.
|
|
52772
|
ADP glucose pyrophosphorylase from maize endosperm.
|
|
52773
|
ADP glucose pyrophosphorylase from maize endosperm.
|
|
52774
|
Construction and characterization of the chimeric enzymes between the Bacillus subtilis cellulase and an ...
|
|
52775
|
Construction and characterization of the chimeric enzymes between the Bacillus subtilis cellulase and an ...
|
|
52776
|
Construction and characterization of the chimeric enzymes between the Bacillus subtilis cellulase and an ...
|
|
52777
|
Construction and characterization of the chimeric enzymes between the Bacillus subtilis cellulase and an ...
|
|
52778
|
Restriction of substrate specificity of subtilisin E by introduction of a side chain into a conserved glycine ...
|
|
52779
|
Restriction of substrate specificity of subtilisin E by introduction of a side chain into a conserved glycine ...
|
|
52780
|
Restriction of substrate specificity of subtilisin E by introduction of a side chain into a conserved glycine ...
|
|
52781
|
Restriction of substrate specificity of subtilisin E by introduction of a side chain into a conserved glycine ...
|
|
52782
|
Restriction of substrate specificity of subtilisin E by introduction of a side chain into a conserved glycine ...
|
|
52783
|
Restriction of substrate specificity of subtilisin E by introduction of a side chain into a conserved glycine ...
|
|
52784
|
Restriction of substrate specificity of subtilisin E by introduction of a side chain into a conserved glycine ...
|
|
52785
|
Restriction of substrate specificity of subtilisin E by introduction of a side chain into a conserved glycine ...
|
|
52786
|
Restriction of substrate specificity of subtilisin E by introduction of a side chain into a conserved glycine ...
|
|
52787
|
Restriction of substrate specificity of subtilisin E by introduction of a side chain into a conserved glycine ...
|
|
52788
|
Reduction of the pea ferredoxin-NADP(H) reductase catalytic efficiency by the structuring of a ...
|
|
52789
|
Reduction of the pea ferredoxin-NADP(H) reductase catalytic efficiency by the structuring of a ...
|
|
52790
|
Reduction of the pea ferredoxin-NADP(H) reductase catalytic efficiency by the structuring of a ...
|
|
52791
|
Reduction of the pea ferredoxin-NADP(H) reductase catalytic efficiency by the structuring of a ...
|
|
52792
|
Reduction of the pea ferredoxin-NADP(H) reductase catalytic efficiency by the structuring of a ...
|
|
52793
|
Reduction of the pea ferredoxin-NADP(H) reductase catalytic efficiency by the structuring of a ...
|
|
52794
|
Reduction of the pea ferredoxin-NADP(H) reductase catalytic efficiency by the structuring of a ...
|
|
52795
|
Reduction of the pea ferredoxin-NADP(H) reductase catalytic efficiency by the structuring of a ...
|
|
52796
|
Reduction of the pea ferredoxin-NADP(H) reductase catalytic efficiency by the structuring of a ...
|
|
52797
|
Preparation and Properties of a beta-d-Glucanase for the Specific Hydrolysis of beta-d-Glucans
|
|
52798
|
Properties of uridine diphosphoglucose dehydrogenase from pollen of Lilium longiflorum
|
|
52799
|
Properties of uridine diphosphoglucose dehydrogenase from pollen of Lilium longiflorum
|
|
52800
|
Properties of uridine diphosphoglucose dehydrogenase from pollen of Lilium longiflorum
|
|
52801
|
Properties of uridine diphosphoglucose dehydrogenase from pollen of Lilium longiflorum
|
|
52802
|
Properties of uridine diphosphoglucose dehydrogenase from pollen of Lilium longiflorum
|
|
52803
|
Properties of uridine diphosphoglucose dehydrogenase from pollen of Lilium longiflorum
|
|
52804
|
Properties of uridine diphosphoglucose dehydrogenase from pollen of Lilium longiflorum
|
|
52805
|
Properties of uridine diphosphoglucose dehydrogenase from pollen of Lilium longiflorum
|
|
52806
|
A comparison of kinetic parameters obtained with three major non-interconvertible isozymes of rat pyruvate ...
|
|
52807
|
A comparison of kinetic parameters obtained with three major non-interconvertible isozymes of rat pyruvate ...
|
|
52808
|
A comparison of kinetic parameters obtained with three major non-interconvertible isozymes of rat pyruvate ...
|
|
52809
|
A comparison of kinetic parameters obtained with three major non-interconvertible isozymes of rat pyruvate ...
|
|
52810
|
A comparison of kinetic parameters obtained with three major non-interconvertible isozymes of rat pyruvate ...
|
|
52811
|
A comparison of kinetic parameters obtained with three major non-interconvertible isozymes of rat pyruvate ...
|
|
52812
|
A comparison of kinetic parameters obtained with three major non-interconvertible isozymes of rat pyruvate ...
|
|
52813
|
A comparison of kinetic parameters obtained with three major non-interconvertible isozymes of rat pyruvate ...
|
|
52814
|
A comparison of kinetic parameters obtained with three major non-interconvertible isozymes of rat pyruvate ...
|
|
52815
|
A comparison of kinetic parameters obtained with three major non-interconvertible isozymes of rat pyruvate ...
|
|
52816
|
A comparison of kinetic parameters obtained with three major non-interconvertible isozymes of rat pyruvate ...
|
|
52817
|
A comparison of kinetic parameters obtained with three major non-interconvertible isozymes of rat pyruvate ...
|
|
52818
|
A comparison of kinetic parameters obtained with three major non-interconvertible isozymes of rat pyruvate ...
|
|
52819
|
A comparison of kinetic parameters obtained with three major non-interconvertible isozymes of rat pyruvate ...
|
|
52820
|
Inhibition of human steroid 5beta-reductase (AKR1D1) by finasteride and structure of the enzyme-inhibitor ...
|
|
52821
|
Inhibition of human steroid 5beta-reductase (AKR1D1) by finasteride and structure of the enzyme-inhibitor ...
|
|
52822
|
Determination of the kinetic parameters for phospholipase C (Bacillus cereus) on different phospholipid ...
|
|
52823
|
Determination of the kinetic parameters for phospholipase C (Bacillus cereus) on different phospholipid ...
|
|
52824
|
Determination of the kinetic parameters for phospholipase C (Bacillus cereus) on different phospholipid ...
|
|
52825
|
Purification and properties of phosphoribosyl-diphosphate synthetase from Bacillus subtilis
|
|
52826
|
Purification and properties of phosphoribosyl-diphosphate synthetase from Bacillus subtilis
|
|
52827
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52828
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52829
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52830
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52831
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52832
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52833
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52834
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52835
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52836
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52837
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52838
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52839
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52840
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52841
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52842
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52843
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52844
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52845
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52846
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52847
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52848
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52849
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52850
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52851
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52852
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52853
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52854
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52855
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52856
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52857
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52858
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52859
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52860
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52861
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52862
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52863
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52864
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52865
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52866
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52867
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52868
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52869
|
Escherichia coli tryptophanyl-tRNA synthetase mutants selected for tryptophan auxotrophy implicate the dimer ...
|
|
52870
|
Kinetic studies on the prenyl chain elongation by undecaprenyl diphosphate synthase with artificial substrate ...
|
|
52871
|
Kinetic studies on the prenyl chain elongation by undecaprenyl diphosphate synthase with artificial substrate ...
|
|
52872
|
Kinetic studies on the prenyl chain elongation by undecaprenyl diphosphate synthase with artificial substrate ...
|
|
52873
|
Kinetic studies on the prenyl chain elongation by undecaprenyl diphosphate synthase with artificial substrate ...
|
|
52874
|
Kinetic studies on the prenyl chain elongation by undecaprenyl diphosphate synthase with artificial substrate ...
|
|
52875
|
Kinetic studies on the prenyl chain elongation by undecaprenyl diphosphate synthase with artificial substrate ...
|
|
52876
|
Overexpression and purification of the trimeric aspartate transcarbamoylase from Bacillus subtilis
|
|
52877
|
Overexpression and purification of the trimeric aspartate transcarbamoylase from Bacillus subtilis
|
|
52878
|
Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate
|
|
52879
|
Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate
|
|
52880
|
Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate
|
|
52881
|
Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate
|
|
52882
|
Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate
|
|
52883
|
Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate
|
|
52884
|
Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate
|
|
52885
|
Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate
|
|
52886
|
Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate
|
|
52887
|
Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate
|
|
52888
|
Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate
|
|
52889
|
Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate
|
|
52890
|
Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate
|
|
52891
|
Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate
|
|
52892
|
Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate
|
|
52893
|
Grafting of a calcium-binding loop of thermolysin to Bacillus subtilis neutral protease
|
|
52894
|
Grafting of a calcium-binding loop of thermolysin to Bacillus subtilis neutral protease
|
|
52895
|
Grafting of a calcium-binding loop of thermolysin to Bacillus subtilis neutral protease
|
|
52896
|
Monofunctional Chorismate Mutase from Bacillus subtilis: Purification of the Protein, Molecular Cloning of the ...
|
|
52897
|
Monofunctional chorismate mutase from Bacillus subtilis: kinetic and 13C NMR studies on the interactions of ...
|
|
52898
|
Monofunctional chorismate mutase from Bacillus subtilis: kinetic and 13C NMR studies on the interactions of ...
|
|
52899
|
Mono- and dimethylating activities and kinetic studies of the ermC 23 S rRNA methyltransferase
|
|
52900
|
Mono- and dimethylating activities and kinetic studies of the ermC 23 S rRNA methyltransferase
|
|
52901
|
Mono- and dimethylating activities and kinetic studies of the ermC 23 S rRNA methyltransferase
|
|
52902
|
Mono- and dimethylating activities and kinetic studies of the ermC 23 S rRNA methyltransferase
|
|
52903
|
Mono- and dimethylating activities and kinetic studies of the ermC 23 S rRNA methyltransferase
|
|
52904
|
Mono- and dimethylating activities and kinetic studies of the ermC 23 S rRNA methyltransferase
|
|
52905
|
Role of the protein moiety of ribonuclease P, a ribonucleoprotein enzyme
|
|
52906
|
Role of the protein moiety of ribonuclease P, a ribonucleoprotein enzyme
|
|
52907
|
Role of the protein moiety of ribonuclease P, a ribonucleoprotein enzyme
|
|
52908
|
Role of the protein moiety of ribonuclease P, a ribonucleoprotein enzyme
|
|
52909
|
Role of the protein moiety of ribonuclease P, a ribonucleoprotein enzyme
|
|
52910
|
Prephenate dehydratase of the actinomycete Amycolatopsis methanolica: purification and characterization of ...
|
|
52911
|
Prephenate dehydratase of the actinomycete Amycolatopsis methanolica: purification and characterization of ...
|
|
52912
|
Prephenate dehydratase of the actinomycete Amycolatopsis methanolica: purification and characterization of ...
|
|
52913
|
Prephenate dehydratase of the actinomycete Amycolatopsis methanolica: purification and characterization of ...
|
|
52914
|
Prephenate dehydratase of the actinomycete Amycolatopsis methanolica: purification and characterization of ...
|
|
52915
|
Prephenate dehydratase of the actinomycete Amycolatopsis methanolica: purification and characterization of ...
|
|
52916
|
Prephenate dehydratase of the actinomycete Amycolatopsis methanolica: purification and characterization of ...
|
|
52917
|
Prephenate dehydratase of the actinomycete Amycolatopsis methanolica: purification and characterization of ...
|
|
52918
|
Prephenate dehydratase of the actinomycete Amycolatopsis methanolica: purification and characterization of ...
|
|
52919
|
Prephenate dehydratase of the actinomycete Amycolatopsis methanolica: purification and characterization of ...
|
|
52920
|
Prephenate dehydratase of the actinomycete Amycolatopsis methanolica: purification and characterization of ...
|
|
52921
|
Prephenate dehydratase of the actinomycete Amycolatopsis methanolica: purification and characterization of ...
|
|
52922
|
Prephenate dehydratase of the actinomycete Amycolatopsis methanolica: purification and characterization of ...
|
|
52923
|
Prephenate dehydratase of the actinomycete Amycolatopsis methanolica: purification and characterization of ...
|
|
52924
|
Properties of serine:glyoxylate aminotransferase purified from Arabidopsis thaliana leaves
|
|
52925
|
Properties of serine:glyoxylate aminotransferase purified from Arabidopsis thaliana leaves
|
|
52926
|
Properties of serine:glyoxylate aminotransferase purified from Arabidopsis thaliana leaves
|
|
52927
|
Properties of serine:glyoxylate aminotransferase purified from Arabidopsis thaliana leaves
|
|
52928
|
Properties of serine:glyoxylate aminotransferase purified from Arabidopsis thaliana leaves
|
|
52929
|
Properties of serine:glyoxylate aminotransferase purified from Arabidopsis thaliana leaves
|
|
52930
|
Properties of serine:glyoxylate aminotransferase purified from Arabidopsis thaliana leaves
|
|
52931
|
Properties of serine:glyoxylate aminotransferase purified from Arabidopsis thaliana leaves
|
|
52932
|
Properties of serine:glyoxylate aminotransferase purified from Arabidopsis thaliana leaves
|
|
52933
|
Properties of serine:glyoxylate aminotransferase purified from Arabidopsis thaliana leaves
|
|
52934
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52935
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52936
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52937
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52938
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52939
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52940
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52941
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52942
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52943
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52944
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52945
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52946
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52947
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52948
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52949
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52950
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52951
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52952
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52953
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52954
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52955
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52956
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52957
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52958
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52959
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52960
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52961
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52962
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52963
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52964
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52965
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52966
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52967
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52968
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52969
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52970
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52971
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52972
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52973
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52974
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52975
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52976
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52977
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52978
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52979
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52980
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52981
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52982
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52983
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52984
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52985
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52986
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52987
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52988
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52989
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52990
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52991
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52992
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52993
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52994
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52995
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52996
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52997
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52998
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
52999
|
Aminofurazans as potent inhibitors of AKT kinase
|
|
53000
|
Aminofurazans as potent inhibitors of AKT kinase
|





 If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.
If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.