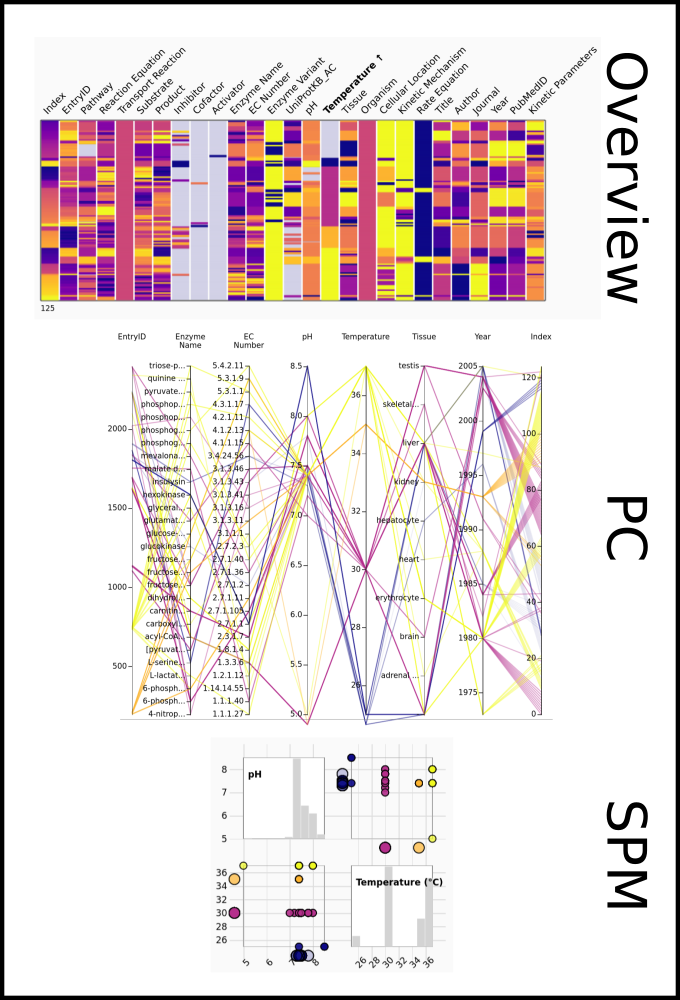
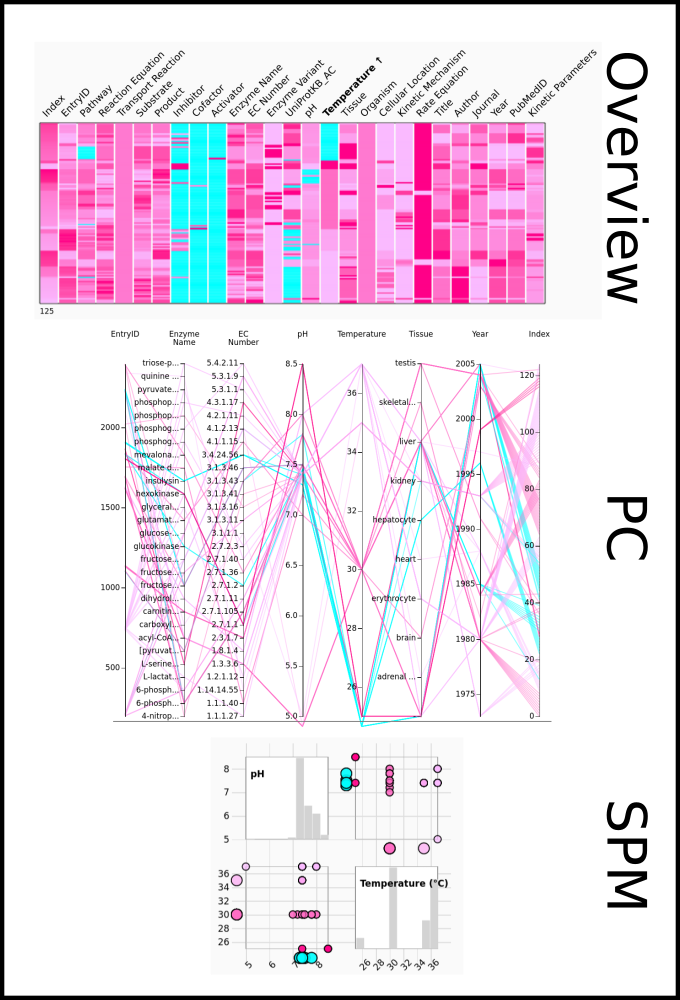
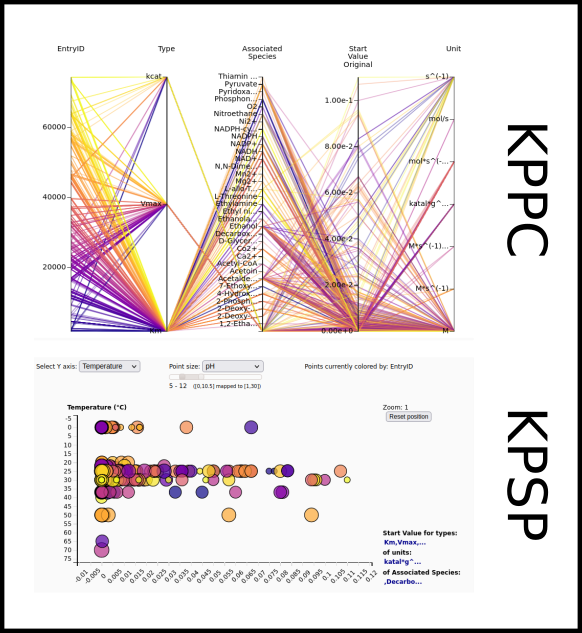
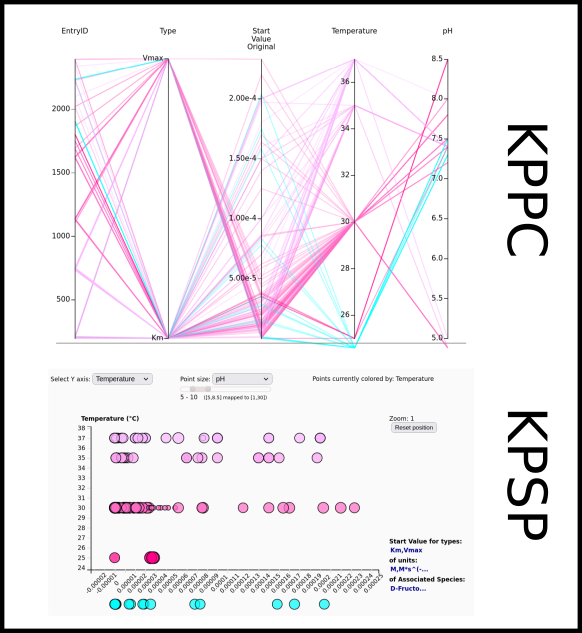
To improve SABIO-RK data interoperability, semantic markup was added to web pages as described and defined by
This structured information makes it easier to discover, collate and analyse our data.
|
42001
|
Identification of amino acid residues critical for catalysis and cosubstrate binding in the flavonol ...
|
|
42002
|
Identification of amino acid residues critical for catalysis and cosubstrate binding in the flavonol ...
|
|
42003
|
Identification of amino acid residues critical for catalysis and cosubstrate binding in the flavonol ...
|
|
42004
|
Identification of amino acid residues critical for catalysis and cosubstrate binding in the flavonol ...
|
|
42005
|
Identification of amino acid residues critical for catalysis and cosubstrate binding in the flavonol ...
|
|
42006
|
Identification of amino acid residues critical for catalysis and cosubstrate binding in the flavonol ...
|
|
42007
|
Identification of amino acid residues critical for catalysis and cosubstrate binding in the flavonol ...
|
|
42008
|
Identification of amino acid residues critical for catalysis and cosubstrate binding in the flavonol ...
|
|
42009
|
Identification of amino acid residues critical for catalysis and cosubstrate binding in the flavonol ...
|
|
42010
|
Identification of amino acid residues critical for catalysis and cosubstrate binding in the flavonol ...
|
|
42011
|
Identification of amino acid residues critical for catalysis and cosubstrate binding in the flavonol ...
|
|
42012
|
Identification of amino acid residues critical for catalysis and cosubstrate binding in the flavonol ...
|
|
42013
|
Characterization and mutagenesis of Gal/GlcNAc-6-O-sulfotransferases
|
|
42014
|
Characterization and mutagenesis of Gal/GlcNAc-6-O-sulfotransferases
|
|
42015
|
Characterization and mutagenesis of Gal/GlcNAc-6-O-sulfotransferases
|
|
42016
|
Characterization and mutagenesis of Gal/GlcNAc-6-O-sulfotransferases
|
|
42017
|
Characterization and mutagenesis of Gal/GlcNAc-6-O-sulfotransferases
|
|
42018
|
Characterization and mutagenesis of Gal/GlcNAc-6-O-sulfotransferases
|
|
42019
|
Characterization and mutagenesis of Gal/GlcNAc-6-O-sulfotransferases
|
|
42020
|
Characterization and mutagenesis of Gal/GlcNAc-6-O-sulfotransferases
|
|
42021
|
Characterization and mutagenesis of Gal/GlcNAc-6-O-sulfotransferases
|
|
42022
|
Characterization and mutagenesis of Gal/GlcNAc-6-O-sulfotransferases
|
|
42023
|
Characterization and mutagenesis of Gal/GlcNAc-6-O-sulfotransferases
|
|
42024
|
Characterization and mutagenesis of Gal/GlcNAc-6-O-sulfotransferases
|
|
42025
|
The probiotic Lactobacillus johnsonii NCC 533 produces high-molecular-mass inulin from sucrose by using an ...
|
|
42026
|
The probiotic Lactobacillus johnsonii NCC 533 produces high-molecular-mass inulin from sucrose by using an ...
|
|
42027
|
The probiotic Lactobacillus johnsonii NCC 533 produces high-molecular-mass inulin from sucrose by using an ...
|
|
42028
|
Phosphatidylethanolamine synthesis by castor bean endosperm. Intracellular distribution and characteristics of ...
|
|
42029
|
Phosphatidylethanolamine synthesis by castor bean endosperm. Intracellular distribution and characteristics of ...
|
|
42030
|
Phosphatidylethanolamine synthesis by castor bean endosperm. Intracellular distribution and characteristics of ...
|
|
42031
|
Phosphatidylethanolamine synthesis by castor bean endosperm. Intracellular distribution and characteristics of ...
|
|
42032
|
Purification, structure, and catalytic properties of L-myo-inositol-1-phosphate synthase from rat testis
|
|
42033
|
Purification, structure, and catalytic properties of L-myo-inositol-1-phosphate synthase from rat testis
|
|
42034
|
Purification, structure, and catalytic properties of L-myo-inositol-1-phosphate synthase from rat testis
|
|
42035
|
Purification, structure, and catalytic properties of L-myo-inositol-1-phosphate synthase from rat testis
|
|
42036
|
Purification, structure, and catalytic properties of L-myo-inositol-1-phosphate synthase from rat testis
|
|
42037
|
Purification, structure, and catalytic properties of L-myo-inositol-1-phosphate synthase from rat testis
|
|
42038
|
Purification, structure, and catalytic properties of L-myo-inositol-1-phosphate synthase from rat testis
|
|
42039
|
Purification, structure, and catalytic properties of L-myo-inositol-1-phosphate synthase from rat testis
|
|
42040
|
Purification, structure, and catalytic properties of L-myo-inositol-1-phosphate synthase from rat testis
|
|
42041
|
Purification, structure, and catalytic properties of L-myo-inositol-1-phosphate synthase from rat testis
|
|
42042
|
Studies on the catalysis of carbon-cobalt bond homolysis by ribonucleoside triphosphate reductase: evidence ...
|
|
42043
|
Studies on the catalysis of carbon-cobalt bond homolysis by ribonucleoside triphosphate reductase: evidence ...
|
|
42044
|
Partial Purification and Characterization of Ribulose-1,5-bisphosphate Carboxylase/Oxygenase Large Subunit ...
|
|
42045
|
Partial Purification and Characterization of Ribulose-1,5-bisphosphate Carboxylase/Oxygenase Large Subunit ...
|
|
42046
|
Purification and characterization of dihydrodipicolinate synthase from pea
|
|
42047
|
Purification and characterization of dihydrodipicolinate synthase from pea
|
|
42048
|
Purification and characterization of dihydrodipicolinate synthase from pea
|
|
42049
|
Purification and characterization of dihydrodipicolinate synthase from pea
|
|
42050
|
Purification and characterization of dihydrodipicolinate synthase from pea
|
|
42051
|
A major urinary protein of the domestic cat regulates the production of felinine, a putative pheromone ...
|
|
42052
|
Purification and characterization of glpX-encoded fructose 1, 6-bisphosphatase, a new enzyme of the glycerol ...
|
|
42053
|
Purification and characterization of glpX-encoded fructose 1, 6-bisphosphatase, a new enzyme of the glycerol ...
|
|
42054
|
Purification and characterization of glpX-encoded fructose 1, 6-bisphosphatase, a new enzyme of the glycerol ...
|
|
42055
|
Purification and characterization of glpX-encoded fructose 1, 6-bisphosphatase, a new enzyme of the glycerol ...
|
|
42056
|
Purification and characterization of glpX-encoded fructose 1, 6-bisphosphatase, a new enzyme of the glycerol ...
|
|
42057
|
The Mycobacterium tuberculosis Rv1099c gene encodes a GlpX-like class II fructose 1,6-bisphosphatase
|
|
42058
|
The Mycobacterium tuberculosis Rv1099c gene encodes a GlpX-like class II fructose 1,6-bisphosphatase
|
|
42059
|
Multisubstrate analogs for deoxynucleoside kinases. Triphosphate end products and synthetic bisubstrate ...
|
|
42060
|
Multisubstrate analogs for deoxynucleoside kinases. Triphosphate end products and synthetic bisubstrate ...
|
|
42061
|
Multisubstrate analogs for deoxynucleoside kinases. Triphosphate end products and synthetic bisubstrate ...
|
|
42062
|
Multisubstrate analogs for deoxynucleoside kinases. Triphosphate end products and synthetic bisubstrate ...
|
|
42063
|
Multisubstrate analogs for deoxynucleoside kinases. Triphosphate end products and synthetic bisubstrate ...
|
|
42064
|
Multisubstrate analogs for deoxynucleoside kinases. Triphosphate end products and synthetic bisubstrate ...
|
|
42065
|
Multisubstrate analogs for deoxynucleoside kinases. Triphosphate end products and synthetic bisubstrate ...
|
|
42066
|
Multisubstrate analogs for deoxynucleoside kinases. Triphosphate end products and synthetic bisubstrate ...
|
|
42067
|
Multisubstrate analogs for deoxynucleoside kinases. Triphosphate end products and synthetic bisubstrate ...
|
|
42068
|
Multisubstrate analogs for deoxynucleoside kinases. Triphosphate end products and synthetic bisubstrate ...
|
|
42069
|
Multisubstrate analogs for deoxynucleoside kinases. Triphosphate end products and synthetic bisubstrate ...
|
|
42070
|
Multisubstrate analogs for deoxynucleoside kinases. Triphosphate end products and synthetic bisubstrate ...
|
|
42071
|
Multisubstrate analogs for deoxynucleoside kinases. Triphosphate end products and synthetic bisubstrate ...
|
|
42072
|
Multisubstrate analogs for deoxynucleoside kinases. Triphosphate end products and synthetic bisubstrate ...
|
|
42073
|
Multisubstrate analogs for deoxynucleoside kinases. Triphosphate end products and synthetic bisubstrate ...
|
|
42074
|
Multisubstrate analogs for deoxynucleoside kinases. Triphosphate end products and synthetic bisubstrate ...
|
|
42075
|
Multisubstrate analogs for deoxynucleoside kinases. Triphosphate end products and synthetic bisubstrate ...
|
|
42076
|
Multisubstrate analogs for deoxynucleoside kinases. Triphosphate end products and synthetic bisubstrate ...
|
|
42077
|
Multisubstrate analogs for deoxynucleoside kinases. Triphosphate end products and synthetic bisubstrate ...
|
|
42078
|
Multisubstrate analogs for deoxynucleoside kinases. Triphosphate end products and synthetic bisubstrate ...
|
|
42079
|
Multisubstrate analogs for deoxynucleoside kinases. Triphosphate end products and synthetic bisubstrate ...
|
|
42080
|
Multisubstrate analogs for deoxynucleoside kinases. Triphosphate end products and synthetic bisubstrate ...
|
|
42081
|
Multisubstrate analogs for deoxynucleoside kinases. Triphosphate end products and synthetic bisubstrate ...
|
|
42082
|
Multisubstrate analogs for deoxynucleoside kinases. Triphosphate end products and synthetic bisubstrate ...
|
|
42083
|
Recombinant N-terminal nucleotide-binding domain from mouse P-glycoprotein. Overexpression, purification, and ...
|
|
42084
|
Conformational change induced by ATP binding in the multidrug ATP-binding cassette transporter BmrA
|
|
42085
|
Conformational change induced by ATP binding in the multidrug ATP-binding cassette transporter BmrA
|
|
42086
|
Conformational change induced by ATP binding in the multidrug ATP-binding cassette transporter BmrA
|
|
42087
|
Conformational change induced by ATP binding in the multidrug ATP-binding cassette transporter BmrA
|
|
42088
|
Conformational change induced by ATP binding in the multidrug ATP-binding cassette transporter BmrA
|
|
42089
|
Conformational change induced by ATP binding in the multidrug ATP-binding cassette transporter BmrA
|
|
42090
|
7,8-Diaminoperlargonic acid aminotransferase from Mycobacterium tuberculosis, a potential therapeutic target. ...
|
|
42091
|
7,8-Diaminoperlargonic acid aminotransferase from Mycobacterium tuberculosis, a potential therapeutic target. ...
|
|
42092
|
7,8-Diaminoperlargonic acid aminotransferase from Mycobacterium tuberculosis, a potential therapeutic target. ...
|
|
42093
|
Novel benzene ring biosynthesis from C(3) and C(4) primary metabolites by two enzymes
|
|
42094
|
Novel benzene ring biosynthesis from C(3) and C(4) primary metabolites by two enzymes
|
|
42095
|
Evidence for intracellular endothelin-converting enzyme-2 expression in cultured human vascular endothelial ...
|
|
42096
|
Isolation and characterization of dihydrodipicolinate synthase from maize
|
|
42097
|
Isolation and characterization of dihydrodipicolinate synthase from maize
|
|
42098
|
Isolation and characterization of dihydrodipicolinate synthase from maize
|
|
42099
|
Isolation and characterization of dihydrodipicolinate synthase from maize
|
|
42100
|
Isolation and characterization of dihydrodipicolinate synthase from maize
|
|
42101
|
Different methylation characteristics of protein arginine methyltransferase 1 and 3 toward the Ewing Sarcoma ...
|
|
42102
|
Different methylation characteristics of protein arginine methyltransferase 1 and 3 toward the Ewing Sarcoma ...
|
|
42103
|
Different methylation characteristics of protein arginine methyltransferase 1 and 3 toward the Ewing Sarcoma ...
|
|
42104
|
Different methylation characteristics of protein arginine methyltransferase 1 and 3 toward the Ewing Sarcoma ...
|
|
42105
|
Identification of proteins interacting with protein arginine methyltransferase 8: the Ewing sarcoma (EWS) ...
|
|
42106
|
Streptococcus salivarius urease: genetic and biochemical characterization and expression in a dental plaque ...
|
|
42107
|
Streptococcus salivarius urease: genetic and biochemical characterization and expression in a dental plaque ...
|
|
42108
|
Streptococcus salivarius urease: genetic and biochemical characterization and expression in a dental plaque ...
|
|
42109
|
Isolation and characterization of cytidine-5'-monophosphate-N-acetylneuraminate hydroxylase from the starfish ...
|
|
42110
|
Isolation and characterization of cytidine-5'-monophosphate-N-acetylneuraminate hydroxylase from the starfish ...
|
|
42111
|
Isolation and characterization of cytidine-5'-monophosphate-N-acetylneuraminate hydroxylase from the starfish ...
|
|
42112
|
Isolation and characterization of cytidine-5'-monophosphate-N-acetylneuraminate hydroxylase from the starfish ...
|
|
42113
|
The biosynthesis of 8-O-methylated sialic acids in the starfish Asterias rubens--isolation and ...
|
|
42114
|
The biosynthesis of 8-O-methylated sialic acids in the starfish Asterias rubens--isolation and ...
|
|
42115
|
The biosynthesis of 8-O-methylated sialic acids in the starfish Asterias rubens--isolation and ...
|
|
42116
|
Purification and characterization of dihydrodipicolinate synthase from wheat suspension cultures
|
|
42117
|
Purification and characterization of dihydrodipicolinate synthase from wheat suspension cultures
|
|
42118
|
Purification and characterization of dihydrodipicolinate synthase from wheat suspension cultures
|
|
42119
|
Purification and characterization of dihydrodipicolinate synthase from wheat suspension cultures
|
|
42120
|
Purification and characterization of dihydrodipicolinate synthase from wheat suspension cultures
|
|
42121
|
Purification and characterization of dihydrodipicolinate synthase from wheat suspension cultures
|
|
42122
|
Purification and characterization of dihydrodipicolinate synthase from wheat suspension cultures
|
|
42123
|
Purification and characterization of dihydrodipicolinate synthase from wheat suspension cultures
|
|
42124
|
Differentially Regulated Isozymes of 3-Deoxy-d-arabino-Heptulosonate-7-Phosphate Synthase from Seedlings of ...
|
|
42125
|
Differentially Regulated Isozymes of 3-Deoxy-d-arabino-Heptulosonate-7-Phosphate Synthase from Seedlings of ...
|
|
42126
|
Differentially Regulated Isozymes of 3-Deoxy-d-arabino-Heptulosonate-7-Phosphate Synthase from Seedlings of ...
|
|
42127
|
The purification of prenyltransferase and isopentenyl pyrophosphate isomerase of pumpkin fruit and their some ...
|
|
42128
|
Biochemical and immunological properties of alkaline invertase isolated from sprouting soybean hypocotyls
|
|
42129
|
Biochemical and immunological properties of alkaline invertase isolated from sprouting soybean hypocotyls
|
|
42130
|
Biochemical and immunological properties of alkaline invertase isolated from sprouting soybean hypocotyls
|
|
42131
|
Biochemical and immunological properties of alkaline invertase isolated from sprouting soybean hypocotyls
|
|
42132
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42133
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42134
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42135
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42136
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42137
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42138
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42139
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42140
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42141
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42142
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42143
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42144
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42145
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42146
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42147
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42148
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42149
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42150
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42151
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42152
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42153
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42154
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42155
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42156
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42157
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42158
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42159
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42160
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42161
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42162
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42163
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42164
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42165
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42166
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42167
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42168
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42169
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42170
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42171
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42172
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42173
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42174
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42175
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42176
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42177
|
Substrate (aglycone) specificity of human cytosolic beta-glucosidase
|
|
42178
|
Purification and characterization of S-adenosyl-L-methionine: norcoclaurine 6-O-methyltransferase from ...
|
|
42179
|
Purification and characterization of S-adenosyl-L-methionine: norcoclaurine 6-O-methyltransferase from ...
|
|
42180
|
Purification and characterization of S-adenosyl-L-methionine: norcoclaurine 6-O-methyltransferase from ...
|
|
42181
|
CpSufE activates the cysteine desulfurase CpNifS for chloroplastic Fe-S cluster formation
|
|
42182
|
CpSufE activates the cysteine desulfurase CpNifS for chloroplastic Fe-S cluster formation
|
|
42183
|
CpSufE activates the cysteine desulfurase CpNifS for chloroplastic Fe-S cluster formation
|
|
42184
|
CpSufE activates the cysteine desulfurase CpNifS for chloroplastic Fe-S cluster formation
|
|
42185
|
Xanthine oxidase in lentil (Lens esculenta) seedlings
|
|
42186
|
Xanthine oxidase in lentil (Lens esculenta) seedlings
|
|
42187
|
Purification and characterization of 1-aminocyclopropane-1-carboxylate synthase from etiolated mung bean ...
|
|
42188
|
Purification and characterization of a phosphoramidon-sensitive endothelin-converting enzyme in porcine aortic ...
|
|
42189
|
Purification and characterization of a phosphoramidon-sensitive endothelin-converting enzyme in porcine aortic ...
|
|
42190
|
Purification and characterization of a phosphoramidon-sensitive endothelin-converting enzyme in porcine aortic ...
|
|
42191
|
Purification and characterization of a phosphoramidon-sensitive endothelin-converting enzyme in porcine aortic ...
|
|
42192
|
Purification and characterization of a phosphoramidon-sensitive endothelin-converting enzyme in porcine aortic ...
|
|
42193
|
Purification and characterization of a phosphoramidon-sensitive endothelin-converting enzyme in porcine aortic ...
|
|
42194
|
Characterization of endothelin-converting enzyme-2. Implication for a role in the nonclassical processing of ...
|
|
42195
|
Characterization of endothelin-converting enzyme-2. Implication for a role in the nonclassical processing of ...
|
|
42196
|
Characterization of endothelin-converting enzyme-2. Implication for a role in the nonclassical processing of ...
|
|
42197
|
Characterization of endothelin-converting enzyme-2. Implication for a role in the nonclassical processing of ...
|
|
42198
|
Characterization of endothelin-converting enzyme-2. Implication for a role in the nonclassical processing of ...
|
|
42199
|
Characterization of a NADH:dichloroindophenol oxidoreductase from Bacillus subtilis
|
|
42200
|
Probing the essential catalytic residues and substrate affinity in the thermoactive Bacillus ...
|
|
42201
|
Probing the essential catalytic residues and substrate affinity in the thermoactive Bacillus ...
|
|
42202
|
Probing the essential catalytic residues and substrate affinity in the thermoactive Bacillus ...
|
|
42203
|
Probing the essential catalytic residues and substrate affinity in the thermoactive Bacillus ...
|
|
42204
|
Probing the essential catalytic residues and substrate affinity in the thermoactive Bacillus ...
|
|
42205
|
Probing the essential catalytic residues and substrate affinity in the thermoactive Bacillus ...
|
|
42206
|
Probing the essential catalytic residues and substrate affinity in the thermoactive Bacillus ...
|
|
42207
|
Probing the essential catalytic residues and substrate affinity in the thermoactive Bacillus ...
|
|
42208
|
The intrachain disulfide bridge is responsible of the unusual stability properties of novel acylphosphatase ...
|
|
42209
|
The intrachain disulfide bridge is responsible of the unusual stability properties of novel acylphosphatase ...
|
|
42210
|
The intrachain disulfide bridge is responsible of the unusual stability properties of novel acylphosphatase ...
|
|
42211
|
The intrachain disulfide bridge is responsible of the unusual stability properties of novel acylphosphatase ...
|
|
42212
|
NAD+-specific D-arabinose dehydrogenase and its contribution to erythroascorbic acid production in ...
|
|
42213
|
NAD+-specific D-arabinose dehydrogenase and its contribution to erythroascorbic acid production in ...
|
|
42214
|
NAD+-specific D-arabinose dehydrogenase and its contribution to erythroascorbic acid production in ...
|
|
42215
|
NAD+-specific D-arabinose dehydrogenase and its contribution to erythroascorbic acid production in ...
|
|
42216
|
NAD+-specific D-arabinose dehydrogenase and its contribution to erythroascorbic acid production in ...
|
|
42217
|
NAD+-specific D-arabinose dehydrogenase and its contribution to erythroascorbic acid production in ...
|
|
42218
|
NAD+-specific D-arabinose dehydrogenase and its contribution to erythroascorbic acid production in ...
|
|
42219
|
Characterization of a novel D-lyxose isomerase from Cohnella laevoribosii RI-39 sp. nov
|
|
42220
|
Characterization of a novel D-lyxose isomerase from Cohnella laevoribosii RI-39 sp. nov
|
|
42221
|
Characterization of a novel D-lyxose isomerase from Cohnella laevoribosii RI-39 sp. nov
|
|
42222
|
Molecular cloning, expression and characterization of a novel apoplastic invertase inhibitor from tomato ...
|
|
42223
|
Molecular cloning of N-methylputrescine oxidase from tobacco
|
|
42224
|
Molecular cloning of N-methylputrescine oxidase from tobacco
|
|
42225
|
Molecular cloning of N-methylputrescine oxidase from tobacco
|
|
42226
|
Molecular cloning of N-methylputrescine oxidase from tobacco
|
|
42227
|
Molecular cloning of N-methylputrescine oxidase from tobacco
|
|
42228
|
Molecular cloning of N-methylputrescine oxidase from tobacco
|
|
42229
|
Identification of a peroxisomal acyl-activating enzyme involved in the biosynthesis of jasmonic acid in ...
|
|
42230
|
Identification of a peroxisomal acyl-activating enzyme involved in the biosynthesis of jasmonic acid in ...
|
|
42231
|
Cyclobranol: a substrate for C25-methyl sterol side chains and potent mechanism-based inactivator of plant ...
|
|
42232
|
Cyclobranol: a substrate for C25-methyl sterol side chains and potent mechanism-based inactivator of plant ...
|
|
42233
|
Cyclobranol: a substrate for C25-methyl sterol side chains and potent mechanism-based inactivator of plant ...
|
|
42234
|
Cyclobranol: a substrate for C25-methyl sterol side chains and potent mechanism-based inactivator of plant ...
|
|
42235
|
Diamine Oxidase from Cultured Roots of Hyoscyamus niger: Its Function in Tropane Alkaloid Biosynthesis
|
|
42236
|
Diamine Oxidase from Cultured Roots of Hyoscyamus niger: Its Function in Tropane Alkaloid Biosynthesis
|
|
42237
|
Diamine Oxidase from Cultured Roots of Hyoscyamus niger: Its Function in Tropane Alkaloid Biosynthesis
|
|
42238
|
Diamine Oxidase from Cultured Roots of Hyoscyamus niger: Its Function in Tropane Alkaloid Biosynthesis
|
|
42239
|
Diamine Oxidase from Cultured Roots of Hyoscyamus niger: Its Function in Tropane Alkaloid Biosynthesis
|
|
42240
|
Diamine Oxidase from Cultured Roots of Hyoscyamus niger: Its Function in Tropane Alkaloid Biosynthesis
|
|
42241
|
Diamine Oxidase from Cultured Roots of Hyoscyamus niger: Its Function in Tropane Alkaloid Biosynthesis
|
|
42242
|
Diamine Oxidase from Cultured Roots of Hyoscyamus niger: Its Function in Tropane Alkaloid Biosynthesis
|
|
42243
|
Diamine Oxidase from Cultured Roots of Hyoscyamus niger: Its Function in Tropane Alkaloid Biosynthesis
|
|
42244
|
Phytochelatins, the heavy-metal-binding peptides of plants, are synthesized from glutathione by a specific ...
|
|
42245
|
Phytochelatins, the heavy-metal-binding peptides of plants, are synthesized from glutathione by a specific ...
|
|
42246
|
Purification and characterization of fructose-2,6-bisphosphatase, a substrate-specific cytosolic enzyme from ...
|
|
42247
|
Purification and characterization of fructose-2,6-bisphosphatase, a substrate-specific cytosolic enzyme from ...
|
|
42248
|
Purification and characterization of fructose-2,6-bisphosphatase, a substrate-specific cytosolic enzyme from ...
|
|
42249
|
Purification and characterization of fructose-2,6-bisphosphatase, a substrate-specific cytosolic enzyme from ...
|
|
42250
|
Arabidopsis CYP90B1 catalyses the early C-22 hydroxylation of C27, C28 and C29 sterols
|
|
42251
|
Arabidopsis CYP90B1 catalyses the early C-22 hydroxylation of C27, C28 and C29 sterols
|
|
42252
|
Metabolomics for biotransformations: Intracellular redox cofactor analysis and enzyme kinetics offer insight ...
|
|
42253
|
Metabolomics for biotransformations: Intracellular redox cofactor analysis and enzyme kinetics offer insight ...
|
|
42254
|
Metabolomics for biotransformations: Intracellular redox cofactor analysis and enzyme kinetics offer insight ...
|
|
42255
|
Metabolomics for biotransformations: Intracellular redox cofactor analysis and enzyme kinetics offer insight ...
|
|
42256
|
Metabolomics for biotransformations: Intracellular redox cofactor analysis and enzyme kinetics offer insight ...
|
|
42257
|
Metabolomics for biotransformations: Intracellular redox cofactor analysis and enzyme kinetics offer insight ...
|
|
42258
|
Metabolomics for biotransformations: Intracellular redox cofactor analysis and enzyme kinetics offer insight ...
|
|
42259
|
Metabolomics for biotransformations: Intracellular redox cofactor analysis and enzyme kinetics offer insight ...
|
|
42260
|
Role of the lateral channel in catalase HPII of Escherichia coli
|
|
42261
|
Role of the lateral channel in catalase HPII of Escherichia coli
|
|
42262
|
Role of the lateral channel in catalase HPII of Escherichia coli
|
|
42263
|
Role of the lateral channel in catalase HPII of Escherichia coli
|
|
42264
|
Role of the lateral channel in catalase HPII of Escherichia coli
|
|
42265
|
Role of the lateral channel in catalase HPII of Escherichia coli
|
|
42266
|
Role of the lateral channel in catalase HPII of Escherichia coli
|
|
42267
|
Role of the lateral channel in catalase HPII of Escherichia coli
|
|
42268
|
Role of the lateral channel in catalase HPII of Escherichia coli
|
|
42269
|
Role of the lateral channel in catalase HPII of Escherichia coli
|
|
42270
|
Role of the lateral channel in catalase HPII of Escherichia coli
|
|
42271
|
Role of the lateral channel in catalase HPII of Escherichia coli
|
|
42272
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42273
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42274
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42275
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42276
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42277
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42278
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42279
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42280
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42281
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42282
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42283
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42284
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42285
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42286
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42287
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42288
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42289
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42290
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42291
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42292
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42293
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42294
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42295
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42296
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42297
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42298
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42299
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42300
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42301
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42302
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42303
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42304
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42305
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42306
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42307
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42308
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42309
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42310
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42311
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42312
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42313
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42314
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42315
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42316
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42317
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42318
|
Purification and characterization of NADP+-dependent 3 alpha-hydroxysteroid dehydrogenase from mouse liver ...
|
|
42319
|
Mechanistic analysis of wheat chlorophyllase
|
|
42320
|
Mechanistic analysis of wheat chlorophyllase
|
|
42321
|
Mechanistic analysis of wheat chlorophyllase
|
|
42322
|
Mechanistic analysis of wheat chlorophyllase
|
|
42323
|
Mechanistic analysis of wheat chlorophyllase
|
|
42324
|
Isolation and characterization of enzymes involved in lysine catabolism from sorghum seeds
|
|
42325
|
Isolation and characterization of enzymes involved in lysine catabolism from sorghum seeds
|
|
42326
|
Eugenol and isoeugenol, characteristic aromatic constituents of spices, are biosynthesized via reduction of a ...
|
|
42327
|
Eugenol and isoeugenol, characteristic aromatic constituents of spices, are biosynthesized via reduction of a ...
|
|
42328
|
Inhibition of Bacillus subtilis deoxyribonucleic acid polymerase III by phenylhydrazinopyrimidines. ...
|
|
42329
|
Inhibition of Bacillus subtilis deoxyribonucleic acid polymerase III by phenylhydrazinopyrimidines. ...
|
|
42330
|
Metabolism of 2-oxoaldehydes in yeasts. Purification and characterization of lactaldehyde dehydrogenase from ...
|
|
42331
|
Metabolism of 2-oxoaldehydes in yeasts. Purification and characterization of lactaldehyde dehydrogenase from ...
|
|
42332
|
Metabolism of 2-oxoaldehyde in mold. Purification and characterization of two methylglyoxal reductases from ...
|
|
42333
|
Metabolism of 2-oxoaldehyde in mold. Purification and characterization of two methylglyoxal reductases from ...
|
|
42334
|
Metabolism of 2-oxoaldehyde in mold. Purification and characterization of two methylglyoxal reductases from ...
|
|
42335
|
Metabolism of 2-oxoaldehyde in mold. Purification and characterization of two methylglyoxal reductases from ...
|
|
42336
|
Metabolism of 2-oxoaldehyde in mold. Purification and characterization of two methylglyoxal reductases from ...
|
|
42337
|
Metabolism of 2-oxoaldehyde in mold. Purification and characterization of two methylglyoxal reductases from ...
|
|
42338
|
Metabolism of 2-oxoaldehyde in mold. Purification and characterization of two methylglyoxal reductases from ...
|
|
42339
|
Metabolism of 2-oxoaldehyde in mold. Purification and characterization of two methylglyoxal reductases from ...
|
|
42340
|
Characterization of two fructosyl-amino acid oxidase homologs of Schizosaccharomyces pombe
|
|
42341
|
Characterization of two fructosyl-amino acid oxidase homologs of Schizosaccharomyces pombe
|
|
42342
|
Characterization of two fructosyl-amino acid oxidase homologs of Schizosaccharomyces pombe
|
|
42343
|
Elucidation of human choline kinase crystal structures in complex with the products ADP or phosphocholine
|
|
42344
|
Elucidation of human choline kinase crystal structures in complex with the products ADP or phosphocholine
|
|
42345
|
Elucidation of human choline kinase crystal structures in complex with the products ADP or phosphocholine
|
|
42346
|
Elucidation of human choline kinase crystal structures in complex with the products ADP or phosphocholine
|
|
42347
|
Elucidation of human choline kinase crystal structures in complex with the products ADP or phosphocholine
|
|
42348
|
Elucidation of human choline kinase crystal structures in complex with the products ADP or phosphocholine
|
|
42349
|
Cloning and characterization of a Nicotiana tabacum methylputrescine oxidase transcript
|
|
42350
|
Cloning and characterization of a Nicotiana tabacum methylputrescine oxidase transcript
|
|
42351
|
Cloning and characterization of a Nicotiana tabacum methylputrescine oxidase transcript
|
|
42352
|
Cloning and characterization of a Nicotiana tabacum methylputrescine oxidase transcript
|
|
42353
|
(R,S)-Reticuline 7-O-methyltransferase and (R,S)-norcoclaurine 6-O-methyltransferase of Papaver somniferum - ...
|
|
42354
|
(R,S)-Reticuline 7-O-methyltransferase and (R,S)-norcoclaurine 6-O-methyltransferase of Papaver somniferum - ...
|
|
42355
|
(R,S)-Reticuline 7-O-methyltransferase and (R,S)-norcoclaurine 6-O-methyltransferase of Papaver somniferum - ...
|
|
42356
|
(R,S)-Reticuline 7-O-methyltransferase and (R,S)-norcoclaurine 6-O-methyltransferase of Papaver somniferum - ...
|
|
42357
|
(R,S)-Reticuline 7-O-methyltransferase and (R,S)-norcoclaurine 6-O-methyltransferase of Papaver somniferum - ...
|
|
42358
|
(R,S)-Reticuline 7-O-methyltransferase and (R,S)-norcoclaurine 6-O-methyltransferase of Papaver somniferum - ...
|
|
42359
|
(R,S)-Reticuline 7-O-methyltransferase and (R,S)-norcoclaurine 6-O-methyltransferase of Papaver somniferum - ...
|
|
42360
|
(R,S)-Reticuline 7-O-methyltransferase and (R,S)-norcoclaurine 6-O-methyltransferase of Papaver somniferum - ...
|
|
42361
|
(R,S)-Reticuline 7-O-methyltransferase and (R,S)-norcoclaurine 6-O-methyltransferase of Papaver somniferum - ...
|
|
42362
|
(R,S)-Reticuline 7-O-methyltransferase and (R,S)-norcoclaurine 6-O-methyltransferase of Papaver somniferum - ...
|
|
42363
|
Barley (Hordeum vulgare) oxalate oxidase is a manganese-containing enzyme
|
|
42364
|
Lysine-ketoglutarate reductase activity in developing maize endosperm
|
|
42365
|
Lysine-ketoglutarate reductase activity in developing maize endosperm
|
|
42366
|
Hyoscyamine 6beta-hydroxylase, a 2-oxoglutarate-dependent dioxygenase, in alkaloid-producing root cultures
|
|
42367
|
Hyoscyamine 6beta-hydroxylase, a 2-oxoglutarate-dependent dioxygenase, in alkaloid-producing root cultures
|
|
42368
|
Hyoscyamine 6beta-hydroxylase, a 2-oxoglutarate-dependent dioxygenase, in alkaloid-producing root cultures
|
|
42369
|
Barley malt-alpha-amylase. Purification, action pattern, and subsite mapping of isozyme 1 and two members of ...
|
|
42370
|
Barley malt-alpha-amylase. Purification, action pattern, and subsite mapping of isozyme 1 and two members of ...
|
|
42371
|
Barley malt-alpha-amylase. Purification, action pattern, and subsite mapping of isozyme 1 and two members of ...
|
|
42372
|
Barley malt-alpha-amylase. Purification, action pattern, and subsite mapping of isozyme 1 and two members of ...
|
|
42373
|
Barley malt-alpha-amylase. Purification, action pattern, and subsite mapping of isozyme 1 and two members of ...
|
|
42374
|
Barley malt-alpha-amylase. Purification, action pattern, and subsite mapping of isozyme 1 and two members of ...
|
|
42375
|
Barley malt-alpha-amylase. Purification, action pattern, and subsite mapping of isozyme 1 and two members of ...
|
|
42376
|
Barley malt-alpha-amylase. Purification, action pattern, and subsite mapping of isozyme 1 and two members of ...
|
|
42377
|
Barley malt-alpha-amylase. Purification, action pattern, and subsite mapping of isozyme 1 and two members of ...
|
|
42378
|
Barley malt-alpha-amylase. Purification, action pattern, and subsite mapping of isozyme 1 and two members of ...
|
|
42379
|
Barley malt-alpha-amylase. Purification, action pattern, and subsite mapping of isozyme 1 and two members of ...
|
|
42380
|
Barley malt-alpha-amylase. Purification, action pattern, and subsite mapping of isozyme 1 and two members of ...
|
|
42381
|
Barley malt-alpha-amylase. Purification, action pattern, and subsite mapping of isozyme 1 and two members of ...
|
|
42382
|
Barley malt-alpha-amylase. Purification, action pattern, and subsite mapping of isozyme 1 and two members of ...
|
|
42383
|
Barley malt-alpha-amylase. Purification, action pattern, and subsite mapping of isozyme 1 and two members of ...
|
|
42384
|
Barley malt-alpha-amylase. Purification, action pattern, and subsite mapping of isozyme 1 and two members of ...
|
|
42385
|
Barley malt-alpha-amylase. Purification, action pattern, and subsite mapping of isozyme 1 and two members of ...
|
|
42386
|
Barley malt-alpha-amylase. Purification, action pattern, and subsite mapping of isozyme 1 and two members of ...
|
|
42387
|
Barley malt-alpha-amylase. Purification, action pattern, and subsite mapping of isozyme 1 and two members of ...
|
|
42388
|
Characterization of the type I dehydroquinase from Salmonella typhi
|
|
42389
|
Activation of pyruvate kinase of Mucor Rouxii by manganese ions
|
|
42390
|
Activation of pyruvate kinase of Mucor Rouxii by manganese ions
|
|
42391
|
Activation of pyruvate kinase of Mucor Rouxii by manganese ions
|
|
42392
|
Activation of pyruvate kinase of Mucor Rouxii by manganese ions
|
|
42393
|
Activation of pyruvate kinase of Mucor Rouxii by manganese ions
|
|
42394
|
Analysis of phosphofructokinase subunits and isozymes in ascites tumor cells and its original tissue, murine ...
|
|
42395
|
Analysis of phosphofructokinase subunits and isozymes in ascites tumor cells and its original tissue, murine ...
|
|
42396
|
Analysis of phosphofructokinase subunits and isozymes in ascites tumor cells and its original tissue, murine ...
|
|
42397
|
Enzymatic O-methylation of epinephrine and other catechols
|
|
42398
|
Enzymatic O-methylation of epinephrine and other catechols
|
|
42399
|
Enzymatic O-methylation of epinephrine and other catechols
|
|
42400
|
Characterization of recombinant plant cinnamate 4-hydroxylase produced in yeast. Kinetic and spectral ...
|
|
42401
|
Characterization of recombinant plant cinnamate 4-hydroxylase produced in yeast. Kinetic and spectral ...
|
|
42402
|
On the regulatory properties of the pyruvate kinase from Trypanosoma cruzi epimastigotes
|
|
42403
|
On the regulatory properties of the pyruvate kinase from Trypanosoma cruzi epimastigotes
|
|
42404
|
On the regulatory properties of the pyruvate kinase from Trypanosoma cruzi epimastigotes
|
|
42405
|
On the regulatory properties of the pyruvate kinase from Trypanosoma cruzi epimastigotes
|
|
42406
|
On the regulatory properties of the pyruvate kinase from Trypanosoma cruzi epimastigotes
|
|
42407
|
On the regulatory properties of the pyruvate kinase from Trypanosoma cruzi epimastigotes
|
|
42408
|
On the regulatory properties of the pyruvate kinase from Trypanosoma cruzi epimastigotes
|
|
42409
|
On the regulatory properties of the pyruvate kinase from Trypanosoma cruzi epimastigotes
|
|
42410
|
On the regulatory properties of the pyruvate kinase from Trypanosoma cruzi epimastigotes
|
|
42411
|
On the regulatory properties of the pyruvate kinase from Trypanosoma cruzi epimastigotes
|
|
42412
|
On the regulatory properties of the pyruvate kinase from Trypanosoma cruzi epimastigotes
|
|
42413
|
On the regulatory properties of the pyruvate kinase from Trypanosoma cruzi epimastigotes
|
|
42414
|
On the regulatory properties of the pyruvate kinase from Trypanosoma cruzi epimastigotes
|
|
42415
|
On the regulatory properties of the pyruvate kinase from Trypanosoma cruzi epimastigotes
|
|
42416
|
On the regulatory properties of the pyruvate kinase from Trypanosoma cruzi epimastigotes
|
|
42417
|
Identification and characterization of bacterial cysteine dioxygenases: a new route of cysteine degradation ...
|
|
42418
|
Identification and characterization of bacterial cysteine dioxygenases: a new route of cysteine degradation ...
|
|
42419
|
Identification and characterization of bacterial cysteine dioxygenases: a new route of cysteine degradation ...
|
|
42420
|
Identification and characterization of bacterial cysteine dioxygenases: a new route of cysteine degradation ...
|
|
42421
|
Identification and characterization of bacterial cysteine dioxygenases: a new route of cysteine degradation ...
|
|
42422
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42423
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42424
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42425
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42426
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42427
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42428
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42429
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42430
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42431
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42432
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42433
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42434
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42435
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42436
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42437
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42438
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42439
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42440
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42441
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42442
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42443
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42444
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42445
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42446
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42447
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42448
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42449
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42450
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42451
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42452
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42453
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42454
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42455
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42456
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42457
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42458
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42459
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42460
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42461
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42462
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42463
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42464
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42465
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42466
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42467
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42468
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42469
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42470
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42471
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42472
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42473
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42474
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42475
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42476
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42477
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42478
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42479
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42480
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42481
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42482
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42483
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42484
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42485
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42486
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42487
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42488
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42489
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42490
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42491
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42492
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42493
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42494
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42495
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42496
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42497
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42498
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42499
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42500
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42501
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42502
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42503
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42504
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42505
|
Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase
|
|
42506
|
Fructose-1,6-bisphosphate-activated pyruvate kinase from Escherichia coli. Nature of bonds involved in the ...
|
|
42507
|
Fructose-1,6-bisphosphate-activated pyruvate kinase from Escherichia coli. Nature of bonds involved in the ...
|
|
42508
|
Fructose-1,6-bisphosphate-activated pyruvate kinase from Escherichia coli. Nature of bonds involved in the ...
|
|
42509
|
Fructose-1,6-bisphosphate-activated pyruvate kinase from Escherichia coli. Nature of bonds involved in the ...
|
|
42510
|
Fructose-1,6-bisphosphate-activated pyruvate kinase from Escherichia coli. Nature of bonds involved in the ...
|
|
42511
|
Fructose-1,6-bisphosphate-activated pyruvate kinase from Escherichia coli. Nature of bonds involved in the ...
|
|
42512
|
Fructose-1,6-bisphosphate-activated pyruvate kinase from Escherichia coli. Nature of bonds involved in the ...
|
|
42513
|
Fructose-1,6-bisphosphate-activated pyruvate kinase from Escherichia coli. Nature of bonds involved in the ...
|
|
42514
|
Fructose-1,6-bisphosphate-activated pyruvate kinase from Escherichia coli. Nature of bonds involved in the ...
|
|
42515
|
Fructose-1,6-bisphosphate-activated pyruvate kinase from Escherichia coli. Nature of bonds involved in the ...
|
|
42516
|
Fructose-1,6-bisphosphate-activated pyruvate kinase from Escherichia coli. Nature of bonds involved in the ...
|
|
42517
|
Fructose-1,6-bisphosphate-activated pyruvate kinase from Escherichia coli. Nature of bonds involved in the ...
|
|
42518
|
Fructose-1,6-bisphosphate-activated pyruvate kinase from Escherichia coli. Nature of bonds involved in the ...
|
|
42519
|
Fructose-1,6-bisphosphate-activated pyruvate kinase from Escherichia coli. Nature of bonds involved in the ...
|
|
42520
|
Fructose-1,6-bisphosphate-activated pyruvate kinase from Escherichia coli. Nature of bonds involved in the ...
|
|
42521
|
Probing the importance of hydrogen bonds in the active site of the subtilisin nattokinase by site-directed ...
|
|
42522
|
Probing the importance of hydrogen bonds in the active site of the subtilisin nattokinase by site-directed ...
|
|
42523
|
Probing the importance of hydrogen bonds in the active site of the subtilisin nattokinase by site-directed ...
|
|
42524
|
Probing the importance of hydrogen bonds in the active site of the subtilisin nattokinase by site-directed ...
|
|
42525
|
Probing the importance of hydrogen bonds in the active site of the subtilisin nattokinase by site-directed ...
|
|
42526
|
Enzymology of l-Tyrosine Biosynthesis in Mung Bean (Vigna radiata [L.] Wilczek)
|
|
42527
|
Enzymology of l-Tyrosine Biosynthesis in Mung Bean (Vigna radiata [L.] Wilczek)
|
|
42528
|
Enzymology of l-Tyrosine Biosynthesis in Mung Bean (Vigna radiata [L.] Wilczek)
|
|
42529
|
Enzymology of l-Tyrosine Biosynthesis in Mung Bean (Vigna radiata [L.] Wilczek)
|
|
42530
|
Enzymology of l-Tyrosine Biosynthesis in Mung Bean (Vigna radiata [L.] Wilczek)
|
|
42531
|
Enzymology of l-Tyrosine Biosynthesis in Mung Bean (Vigna radiata [L.] Wilczek)
|
|
42532
|
Purification of the ATPase of Streptococcus faecalis
|
|
42533
|
Purification of the ATPase of Streptococcus faecalis
|
|
42534
|
Molecular and immunological characterization of plastid and cytosolic pyruvate kinase isozymes from ...
|
|
42535
|
Molecular and immunological characterization of plastid and cytosolic pyruvate kinase isozymes from ...
|
|
42536
|
Molecular and immunological characterization of plastid and cytosolic pyruvate kinase isozymes from ...
|
|
42537
|
Molecular and immunological characterization of plastid and cytosolic pyruvate kinase isozymes from ...
|
|
42538
|
lac Repressor-operator interaction. VI. The natural inducer of the lac operon
|
|
42539
|
myo-Inositol-1-phosphate synthase from pine pollen: sulfhydryl involvement at the active site
|
|
42540
|
Dual enzymatic routes to L-tyrosine and L-phenylalanine via pretyrosine in Pseudomonas aeruginosa
|
|
42541
|
Dual enzymatic routes to L-tyrosine and L-phenylalanine via pretyrosine in Pseudomonas aeruginosa
|
|
42542
|
Dual enzymatic routes to L-tyrosine and L-phenylalanine via pretyrosine in Pseudomonas aeruginosa
|
|
42543
|
Dual enzymatic routes to L-tyrosine and L-phenylalanine via pretyrosine in Pseudomonas aeruginosa
|
|
42544
|
Dual enzymatic routes to L-tyrosine and L-phenylalanine via pretyrosine in Pseudomonas aeruginosa
|
|
42545
|
Dual enzymatic routes to L-tyrosine and L-phenylalanine via pretyrosine in Pseudomonas aeruginosa
|
|
42546
|
Dual enzymatic routes to L-tyrosine and L-phenylalanine via pretyrosine in Pseudomonas aeruginosa
|
|
42547
|
Dual enzymatic routes to L-tyrosine and L-phenylalanine via pretyrosine in Pseudomonas aeruginosa
|
|
42548
|
Dual enzymatic routes to L-tyrosine and L-phenylalanine via pretyrosine in Pseudomonas aeruginosa
|
|
42549
|
Dual enzymatic routes to L-tyrosine and L-phenylalanine via pretyrosine in Pseudomonas aeruginosa
|
|
42550
|
Dual enzymatic routes to L-tyrosine and L-phenylalanine via pretyrosine in Pseudomonas aeruginosa
|
|
42551
|
Identification and functional analysis of common human flavin-containing monooxygenase 3 genetic variants
|
|
42552
|
Identification and functional analysis of common human flavin-containing monooxygenase 3 genetic variants
|
|
42553
|
Identification and functional analysis of common human flavin-containing monooxygenase 3 genetic variants
|
|
42554
|
Identification and functional analysis of common human flavin-containing monooxygenase 3 genetic variants
|
|
42555
|
Identification and functional analysis of common human flavin-containing monooxygenase 3 genetic variants
|
|
42556
|
Identification and functional analysis of common human flavin-containing monooxygenase 3 genetic variants
|
|
42557
|
Identification and functional analysis of common human flavin-containing monooxygenase 3 genetic variants
|
|
42558
|
Identification and functional analysis of common human flavin-containing monooxygenase 3 genetic variants
|
|
42559
|
Identification and functional analysis of common human flavin-containing monooxygenase 3 genetic variants
|
|
42560
|
Identification and functional analysis of common human flavin-containing monooxygenase 3 genetic variants
|
|
42561
|
Identification and functional analysis of common human flavin-containing monooxygenase 3 genetic variants
|
|
42562
|
Identification and functional analysis of common human flavin-containing monooxygenase 3 genetic variants
|
|
42563
|
Identification and functional analysis of common human flavin-containing monooxygenase 3 genetic variants
|
|
42564
|
Identification and functional analysis of common human flavin-containing monooxygenase 3 genetic variants
|
|
42565
|
Complete purification and general characterization of FAD synthetase from rat liver
|
|
42566
|
Complete purification and general characterization of FAD synthetase from rat liver
|
|
42567
|
ADPG formation by the ADP-specific cleavage of sucrose-reassessment of sucrose synthase
|
|
42568
|
ADPG formation by the ADP-specific cleavage of sucrose-reassessment of sucrose synthase
|
|
42569
|
ADPG formation by the ADP-specific cleavage of sucrose-reassessment of sucrose synthase
|
|
42570
|
ADPG formation by the ADP-specific cleavage of sucrose-reassessment of sucrose synthase
|
|
42571
|
ADPG formation by the ADP-specific cleavage of sucrose-reassessment of sucrose synthase
|
|
42572
|
ADPG formation by the ADP-specific cleavage of sucrose-reassessment of sucrose synthase
|
|
42573
|
ADPG formation by the ADP-specific cleavage of sucrose-reassessment of sucrose synthase
|
|
42574
|
ADPG formation by the ADP-specific cleavage of sucrose-reassessment of sucrose synthase
|
|
42575
|
Crystal structure of human thioesterase superfamily member 2
|
|
42576
|
Crystal structure of human thioesterase superfamily member 2
|
|
42577
|
Crystal structure of human thioesterase superfamily member 2
|
|
42578
|
Crystal structure of human thioesterase superfamily member 2
|
|
42579
|
Crystal structure of human thioesterase superfamily member 2
|
|
42580
|
Crystal structure of human thioesterase superfamily member 2
|
|
42581
|
Crystal structure of human thioesterase superfamily member 2
|
|
42582
|
Crystal structure of human thioesterase superfamily member 2
|
|
42583
|
Crystal structure of human thioesterase superfamily member 2
|
|
42584
|
Crystal structure of human thioesterase superfamily member 2
|
|
42585
|
Crystal structure of human thioesterase superfamily member 2
|
|
42586
|
Crystal structure of human thioesterase superfamily member 2
|
|
42587
|
Crystal structure of human thioesterase superfamily member 2
|
|
42588
|
Crystal structure of human thioesterase superfamily member 2
|
|
42589
|
Crystal structure of human thioesterase superfamily member 2
|
|
42590
|
Crystal structure of human thioesterase superfamily member 2
|
|
42591
|
Crystal structure of human thioesterase superfamily member 2
|
|
42592
|
Two interconvertible forms of glucose-6-phosphate isomerase in rat muscle
|
|
42593
|
Two interconvertible forms of glucose-6-phosphate isomerase in rat muscle
|
|
42594
|
Two interconvertible forms of glucose-6-phosphate isomerase in rat muscle
|
|
42595
|
Two interconvertible forms of glucose-6-phosphate isomerase in rat muscle
|
|
42596
|
Purification and properties of C 55 -isoprenylpyrophosphate phosphatase from Micrococcus lysodeikticus
|
|
42597
|
The role of cations in avian liver phosphoenolpyruvate carboxykinase catalysis. Activation and regulation
|
|
42598
|
The role of cations in avian liver phosphoenolpyruvate carboxykinase catalysis. Activation and regulation
|
|
42599
|
The role of cations in avian liver phosphoenolpyruvate carboxykinase catalysis. Activation and regulation
|
|
42600
|
The role of cations in avian liver phosphoenolpyruvate carboxykinase catalysis. Activation and regulation
|
|
42601
|
The role of cations in avian liver phosphoenolpyruvate carboxykinase catalysis. Activation and regulation
|
|
42602
|
The role of cations in avian liver phosphoenolpyruvate carboxykinase catalysis. Activation and regulation
|
|
42603
|
The role of cations in avian liver phosphoenolpyruvate carboxykinase catalysis. Activation and regulation
|
|
42604
|
The role of cations in avian liver phosphoenolpyruvate carboxykinase catalysis. Activation and regulation
|
|
42605
|
The role of cations in avian liver phosphoenolpyruvate carboxykinase catalysis. Activation and regulation
|
|
42606
|
The role of cations in avian liver phosphoenolpyruvate carboxykinase catalysis. Activation and regulation
|
|
42607
|
The role of cations in avian liver phosphoenolpyruvate carboxykinase catalysis. Activation and regulation
|
|
42608
|
The role of cations in avian liver phosphoenolpyruvate carboxykinase catalysis. Activation and regulation
|
|
42609
|
The role of cations in avian liver phosphoenolpyruvate carboxykinase catalysis. Activation and regulation
|
|
42610
|
The role of cations in avian liver phosphoenolpyruvate carboxykinase catalysis. Activation and regulation
|
|
42611
|
The role of cations in avian liver phosphoenolpyruvate carboxykinase catalysis. Activation and regulation
|
|
42612
|
The role of cations in avian liver phosphoenolpyruvate carboxykinase catalysis. Activation and regulation
|
|
42613
|
The role of cations in avian liver phosphoenolpyruvate carboxykinase catalysis. Activation and regulation
|
|
42614
|
The role of cations in avian liver phosphoenolpyruvate carboxykinase catalysis. Activation and regulation
|
|
42615
|
Monomeric purine nucleoside phosphorylase from rabbit liver. Purification and characterization
|
|
42616
|
Monomeric purine nucleoside phosphorylase from rabbit liver. Purification and characterization
|
|
42617
|
Monomeric purine nucleoside phosphorylase from rabbit liver. Purification and characterization
|
|
42618
|
Monomeric purine nucleoside phosphorylase from rabbit liver. Purification and characterization
|
|
42619
|
Monomeric purine nucleoside phosphorylase from rabbit liver. Purification and characterization
|
|
42620
|
Monomeric purine nucleoside phosphorylase from rabbit liver. Purification and characterization
|
|
42621
|
Monomeric purine nucleoside phosphorylase from rabbit liver. Purification and characterization
|
|
42622
|
Monomeric purine nucleoside phosphorylase from rabbit liver. Purification and characterization
|
|
42623
|
Monomeric purine nucleoside phosphorylase from rabbit liver. Purification and characterization
|
|
42624
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42625
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42626
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42627
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42628
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42629
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42630
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42631
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42632
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42633
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42634
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42635
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42636
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42637
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42638
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42639
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42640
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42641
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42642
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42643
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42644
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42645
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42646
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42647
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42648
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42649
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42650
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42651
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42652
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42653
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42654
|
Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase
|
|
42655
|
The enzymology of lysine biosynthesis in higher plants. The occurrence, characterization and some regulatory ...
|
|
42656
|
The enzymology of lysine biosynthesis in higher plants. The occurrence, characterization and some regulatory ...
|
|
42657
|
The enzymology of lysine biosynthesis in higher plants. The occurrence, characterization and some regulatory ...
|
|
42658
|
The enzymology of lysine biosynthesis in higher plants. The occurrence, characterization and some regulatory ...
|
|
42659
|
Bifunctional catalysis by CDP-ribitol synthase: convergent recruitment of reductase and cytidylyltransferase ...
|
|
42660
|
Bifunctional catalysis by CDP-ribitol synthase: convergent recruitment of reductase and cytidylyltransferase ...
|
|
42661
|
Bifunctional catalysis by CDP-ribitol synthase: convergent recruitment of reductase and cytidylyltransferase ...
|
|
42662
|
Bifunctional catalysis by CDP-ribitol synthase: convergent recruitment of reductase and cytidylyltransferase ...
|
|
42663
|
Bifunctional catalysis by CDP-ribitol synthase: convergent recruitment of reductase and cytidylyltransferase ...
|
|
42664
|
Bifunctional catalysis by CDP-ribitol synthase: convergent recruitment of reductase and cytidylyltransferase ...
|
|
42665
|
Bifunctional catalysis by CDP-ribitol synthase: convergent recruitment of reductase and cytidylyltransferase ...
|
|
42666
|
Bifunctional catalysis by CDP-ribitol synthase: convergent recruitment of reductase and cytidylyltransferase ...
|
|
42667
|
Bifunctional catalysis by CDP-ribitol synthase: convergent recruitment of reductase and cytidylyltransferase ...
|
|
42668
|
Bifunctional catalysis by CDP-ribitol synthase: convergent recruitment of reductase and cytidylyltransferase ...
|
|
42669
|
Bifunctional catalysis by CDP-ribitol synthase: convergent recruitment of reductase and cytidylyltransferase ...
|
|
42670
|
Bifunctional catalysis by CDP-ribitol synthase: convergent recruitment of reductase and cytidylyltransferase ...
|
|
42671
|
Bifunctional catalysis by CDP-ribitol synthase: convergent recruitment of reductase and cytidylyltransferase ...
|
|
42672
|
Bifunctional catalysis by CDP-ribitol synthase: convergent recruitment of reductase and cytidylyltransferase ...
|
|
42673
|
Bifunctional catalysis by CDP-ribitol synthase: convergent recruitment of reductase and cytidylyltransferase ...
|
|
42674
|
Bifunctional catalysis by CDP-ribitol synthase: convergent recruitment of reductase and cytidylyltransferase ...
|
|
42675
|
Bifunctional catalysis by CDP-ribitol synthase: convergent recruitment of reductase and cytidylyltransferase ...
|
|
42676
|
Bifunctional catalysis by CDP-ribitol synthase: convergent recruitment of reductase and cytidylyltransferase ...
|
|
42677
|
Bifunctional catalysis by CDP-ribitol synthase: convergent recruitment of reductase and cytidylyltransferase ...
|
|
42678
|
Bifunctional catalysis by CDP-ribitol synthase: convergent recruitment of reductase and cytidylyltransferase ...
|
|
42679
|
Purification and substrate specificity of UDP-D-xylose:beta-D-glucoside alpha-1,3-D-xylosyltransferase ...
|
|
42680
|
Purification and substrate specificity of UDP-D-xylose:beta-D-glucoside alpha-1,3-D-xylosyltransferase ...
|
|
42681
|
Purification and Partial Characterization of Potato (Solanum tuberosum) Invertase and Its Endogenous ...
|
|
42682
|
Crystal structure of a type III pantothenate kinase: insight into the mechanism of an essential coenzyme A ...
|
|
42683
|
Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional ...
|
|
42684
|
Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional ...
|
|
42685
|
Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional ...
|
|
42686
|
Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional ...
|
|
42687
|
Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional ...
|
|
42688
|
Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional ...
|
|
42689
|
Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional ...
|
|
42690
|
Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional ...
|
|
42691
|
Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional ...
|
|
42692
|
Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional ...
|
|
42693
|
Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional ...
|
|
42694
|
Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional ...
|
|
42695
|
Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional ...
|
|
42696
|
Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional ...
|
|
42697
|
Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional ...
|
|
42698
|
Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional ...
|
|
42699
|
Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional ...
|
|
42700
|
Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional ...
|
|
42701
|
Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional ...
|
|
42702
|
Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional ...
|
|
42703
|
Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional ...
|
|
42704
|
New role for leucyl aminopeptidase in glutathione turnover
|
|
42705
|
New role for leucyl aminopeptidase in glutathione turnover
|
|
42706
|
New role for leucyl aminopeptidase in glutathione turnover
|
|
42707
|
New role for leucyl aminopeptidase in glutathione turnover
|
|
42708
|
New role for leucyl aminopeptidase in glutathione turnover
|
|
42709
|
New role for leucyl aminopeptidase in glutathione turnover
|
|
42710
|
New role for leucyl aminopeptidase in glutathione turnover
|
|
42711
|
New role for leucyl aminopeptidase in glutathione turnover
|
|
42712
|
New role for leucyl aminopeptidase in glutathione turnover
|
|
42713
|
New role for leucyl aminopeptidase in glutathione turnover
|
|
42714
|
New role for leucyl aminopeptidase in glutathione turnover
|
|
42715
|
Mechanisms of ammonium transport, accumulation, and retention in ooyctes and yeast cells expressing ...
|
|
42716
|
Mechanisms of ammonium transport, accumulation, and retention in ooyctes and yeast cells expressing ...
|
|
42717
|
Expression and characterization of the genes encoding azoreductases from Bacillus subtilis and Geobacillus ...
|
|
42718
|
Expression and characterization of the genes encoding azoreductases from Bacillus subtilis and Geobacillus ...
|
|
42719
|
Expression and characterization of the genes encoding azoreductases from Bacillus subtilis and Geobacillus ...
|
|
42720
|
Expression and characterization of the genes encoding azoreductases from Bacillus subtilis and Geobacillus ...
|
|
42721
|
Expression and characterization of the genes encoding azoreductases from Bacillus subtilis and Geobacillus ...
|
|
42722
|
Expression and characterization of the genes encoding azoreductases from Bacillus subtilis and Geobacillus ...
|
|
42723
|
Biochemical and molecular characterization of an azoreductase from Staphylococcus aureus, a tetrameric ...
|
|
42724
|
Biochemical and molecular characterization of an azoreductase from Staphylococcus aureus, a tetrameric ...
|
|
42725
|
The amino acid sequence of rat liver glucokinase deduced from cloned cDNA
|
|
42726
|
Evidence that NADP+ is the physiological cofactor of ADP-L-glycero-D-mannoheptose 6-epimerase
|
|
42727
|
Evidence that NADP+ is the physiological cofactor of ADP-L-glycero-D-mannoheptose 6-epimerase
|
|
42728
|
Characterization of a thermostable NADPH:FMN oxidoreductase from the mesophilic bacterium Bacillus subtilis
|
|
42729
|
Characterization of a thermostable NADPH:FMN oxidoreductase from the mesophilic bacterium Bacillus subtilis
|
|
42730
|
Characterization of a thermostable NADPH:FMN oxidoreductase from the mesophilic bacterium Bacillus subtilis
|
|
42731
|
Characterization of a thermostable NADPH:FMN oxidoreductase from the mesophilic bacterium Bacillus subtilis
|
|
42732
|
Characterization of a thermostable NADPH:FMN oxidoreductase from the mesophilic bacterium Bacillus subtilis
|
|
42733
|
Characterization of a thermostable NADPH:FMN oxidoreductase from the mesophilic bacterium Bacillus subtilis
|
|
42734
|
Characterization of a beta-N-acetylhexosaminidase and a beta-N-acetylglucosaminidase/beta-glucosidase from ...
|
|
42735
|
Characterization of a beta-N-acetylhexosaminidase and a beta-N-acetylglucosaminidase/beta-glucosidase from ...
|
|
42736
|
Characterization of a beta-N-acetylhexosaminidase and a beta-N-acetylglucosaminidase/beta-glucosidase from ...
|
|
42737
|
Characterization of a beta-N-acetylhexosaminidase and a beta-N-acetylglucosaminidase/beta-glucosidase from ...
|
|
42738
|
Expression, purification, and characterization of a novel G protein, SGP, from Streptococcus mutans
|
|
42739
|
Identification of a novel amidase motif in neutral ceramidase
|
|
42740
|
Topology and active site of PlsY: the bacterial acylphosphate:glycerol-3-phosphate acyltransferase
|
|
42741
|
Topology and active site of PlsY: the bacterial acylphosphate:glycerol-3-phosphate acyltransferase
|
|
42742
|
Topology and active site of PlsY: the bacterial acylphosphate:glycerol-3-phosphate acyltransferase
|
|
42743
|
Topology and active site of PlsY: the bacterial acylphosphate:glycerol-3-phosphate acyltransferase
|
|
42744
|
Topology and active site of PlsY: the bacterial acylphosphate:glycerol-3-phosphate acyltransferase
|
|
42745
|
Topology and active site of PlsY: the bacterial acylphosphate:glycerol-3-phosphate acyltransferase
|
|
42746
|
Topology and active site of PlsY: the bacterial acylphosphate:glycerol-3-phosphate acyltransferase
|
|
42747
|
Fructose-1,6-bisphosphatase from Corynebacterium glutamicum: expression and deletion of the fbp gene and ...
|
|
42748
|
Fructose-1,6-bisphosphatase from Corynebacterium glutamicum: expression and deletion of the fbp gene and ...
|
|
42749
|
Fructose-1,6-bisphosphatase from Corynebacterium glutamicum: expression and deletion of the fbp gene and ...
|
|
42750
|
Fructose-1,6-bisphosphatase from Corynebacterium glutamicum: expression and deletion of the fbp gene and ...
|
|
42751
|
Fructose-1,6-bisphosphatase from Corynebacterium glutamicum: expression and deletion of the fbp gene and ...
|
|
42752
|
Fructose-1,6-bisphosphatase from Corynebacterium glutamicum: expression and deletion of the fbp gene and ...
|
|
42753
|
Fructose-1,6-bisphosphatase from Corynebacterium glutamicum: expression and deletion of the fbp gene and ...
|
|
42754
|
Fructose-1,6-bisphosphatase from Corynebacterium glutamicum: expression and deletion of the fbp gene and ...
|
|
42755
|
Fructose-1,6-bisphosphatase from Corynebacterium glutamicum: expression and deletion of the fbp gene and ...
|
|
42756
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42757
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42758
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42759
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42760
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42761
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42762
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42763
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42764
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42765
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42766
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42767
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42768
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42769
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42770
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42771
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42772
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42773
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42774
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42775
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42776
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42777
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42778
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42779
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42780
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42781
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42782
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42783
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42784
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42785
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42786
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42787
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42788
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42789
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42790
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42791
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42792
|
On the regulation and allosteric model of L-type pyruvate kinase from rat liver
|
|
42793
|
Structural role for Tyr-104 in Escherichia coli isopentenyl-diphosphate isomerase: site-directed mutagenesis, ...
|
|
42794
|
Structural role for Tyr-104 in Escherichia coli isopentenyl-diphosphate isomerase: site-directed mutagenesis, ...
|
|
42795
|
Structural role for Tyr-104 in Escherichia coli isopentenyl-diphosphate isomerase: site-directed mutagenesis, ...
|
|
42796
|
Analysis of two diacylglycerol kinase activities in mixed micelles
|
|
42797
|
Analysis of two diacylglycerol kinase activities in mixed micelles
|
|
42798
|
Analysis of two diacylglycerol kinase activities in mixed micelles
|
|
42799
|
Expression, purification, and characterization of human and rat acetyl coenzyme A carboxylase (ACC) isozymes
|
|
42800
|
Expression, purification, and characterization of human and rat acetyl coenzyme A carboxylase (ACC) isozymes
|
|
42801
|
Expression, purification, and characterization of human and rat acetyl coenzyme A carboxylase (ACC) isozymes
|
|
42802
|
Expression, purification, and characterization of human and rat acetyl coenzyme A carboxylase (ACC) isozymes
|
|
42803
|
Expression, purification, and characterization of human and rat acetyl coenzyme A carboxylase (ACC) isozymes
|
|
42804
|
Expression, purification, and characterization of human and rat acetyl coenzyme A carboxylase (ACC) isozymes
|
|
42805
|
Expression, purification, and characterization of human and rat acetyl coenzyme A carboxylase (ACC) isozymes
|
|
42806
|
Expression, purification, and characterization of human and rat acetyl coenzyme A carboxylase (ACC) isozymes
|
|
42807
|
Expression, purification, and characterization of human and rat acetyl coenzyme A carboxylase (ACC) isozymes
|
|
42808
|
Expression, purification, and characterization of human and rat acetyl coenzyme A carboxylase (ACC) isozymes
|
|
42809
|
Expression, purification, and characterization of human and rat acetyl coenzyme A carboxylase (ACC) isozymes
|
|
42810
|
Adipogenesis-related increase of semicarbazide-sensitive amine oxidase and monoamine oxidase in human ...
|
|
42811
|
Adipogenesis-related increase of semicarbazide-sensitive amine oxidase and monoamine oxidase in human ...
|
|
42812
|
Adipogenesis-related increase of semicarbazide-sensitive amine oxidase and monoamine oxidase in human ...
|
|
42813
|
Adipogenesis-related increase of semicarbazide-sensitive amine oxidase and monoamine oxidase in human ...
|
|
42814
|
Enhancement of riboflavin production with Bacillus subtilis by expression and site-directed mutagenesis of zwf ...
|
|
42815
|
Enhancement of riboflavin production with Bacillus subtilis by expression and site-directed mutagenesis of zwf ...
|
|
42816
|
Enhancement of riboflavin production with Bacillus subtilis by expression and site-directed mutagenesis of zwf ...
|
|
42817
|
Enhancement of riboflavin production with Bacillus subtilis by expression and site-directed mutagenesis of zwf ...
|
|
42818
|
Enhancement of riboflavin production with Bacillus subtilis by expression and site-directed mutagenesis of zwf ...
|
|
42819
|
Enhancement of riboflavin production with Bacillus subtilis by expression and site-directed mutagenesis of zwf ...
|
|
42820
|
Pyruvate kinase from Trichomonas vaginalis, an allosteric enzyme stimulated by ribose 5-phosphate and ...
|
|
42821
|
Pyruvate kinase from Trichomonas vaginalis, an allosteric enzyme stimulated by ribose 5-phosphate and ...
|
|
42822
|
Pyruvate kinase from Trichomonas vaginalis, an allosteric enzyme stimulated by ribose 5-phosphate and ...
|
|
42823
|
Pyruvate kinase from Trichomonas vaginalis, an allosteric enzyme stimulated by ribose 5-phosphate and ...
|
|
42824
|
Pyruvate kinase from Trichomonas vaginalis, an allosteric enzyme stimulated by ribose 5-phosphate and ...
|
|
42825
|
Pyruvate kinase from Trichomonas vaginalis, an allosteric enzyme stimulated by ribose 5-phosphate and ...
|
|
42826
|
Pyruvate kinase from Trichomonas vaginalis, an allosteric enzyme stimulated by ribose 5-phosphate and ...
|
|
42827
|
Pyruvate kinase from Trichomonas vaginalis, an allosteric enzyme stimulated by ribose 5-phosphate and ...
|
|
42828
|
Pyruvate kinase from Trichomonas vaginalis, an allosteric enzyme stimulated by ribose 5-phosphate and ...
|
|
42829
|
Pyruvate kinase from Trichomonas vaginalis, an allosteric enzyme stimulated by ribose 5-phosphate and ...
|
|
42830
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42831
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42832
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42833
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42834
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42835
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42836
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42837
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42838
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42839
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42840
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42841
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42842
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42843
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42844
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42845
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42846
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42847
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42848
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42849
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42850
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42851
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42852
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42853
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42854
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42855
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42856
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42857
|
Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase ...
|
|
42858
|
Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in ...
|
|
42859
|
Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in ...
|
|
42860
|
Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in ...
|
|
42861
|
Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in ...
|
|
42862
|
Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in ...
|
|
42863
|
Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in ...
|
|
42864
|
Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in ...
|
|
42865
|
Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in ...
|
|
42866
|
Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in ...
|
|
42867
|
Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in ...
|
|
42868
|
Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in ...
|
|
42869
|
Additive contribution of AMT1;1 and AMT1;3 to high-affinity ammonium uptake across the plasma membrane of ...
|
|
42870
|
Additive contribution of AMT1;1 and AMT1;3 to high-affinity ammonium uptake across the plasma membrane of ...
|
|
42871
|
Additive contribution of AMT1;1 and AMT1;3 to high-affinity ammonium uptake across the plasma membrane of ...
|
|
42872
|
Additive contribution of AMT1;1 and AMT1;3 to high-affinity ammonium uptake across the plasma membrane of ...
|
|
42873
|
Protective mechanisms against homocysteine toxicity: the role of bleomycin hydrolase
|
|
42874
|
Protective mechanisms against homocysteine toxicity: the role of bleomycin hydrolase
|
|
42875
|
Protective mechanisms against homocysteine toxicity: the role of bleomycin hydrolase
|
|
42876
|
Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell ...
|
|
42877
|
Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell ...
|
|
42878
|
Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell ...
|
|
42879
|
Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell ...
|
|
42880
|
Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell ...
|
|
42881
|
Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell ...
|
|
42882
|
Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell ...
|
|
42883
|
Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell ...
|
|
42884
|
Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell ...
|
|
42885
|
Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell ...
|
|
42886
|
Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell ...
|
|
42887
|
Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell ...
|
|
42888
|
Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell ...
|
|
42889
|
Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell ...
|
|
42890
|
Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell ...
|
|
42891
|
Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell ...
|
|
42892
|
Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell ...
|
|
42893
|
Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell ...
|
|
42894
|
Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell ...
|
|
42895
|
Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell ...
|
|
42896
|
Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell ...
|
|
42897
|
A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis
|
|
42898
|
A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis
|
|
42899
|
A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis
|
|
42900
|
A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis
|
|
42901
|
A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis
|
|
42902
|
A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis
|
|
42903
|
A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis
|
|
42904
|
A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis
|
|
42905
|
A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis
|
|
42906
|
Purification and characterization of pyrophosphate-dependent phosphofructokinase from phosphate-starved ...
|
|
42907
|
Purification and characterization of pyrophosphate-dependent phosphofructokinase from phosphate-starved ...
|
|
42908
|
Purification and characterization of pyrophosphate-dependent phosphofructokinase from phosphate-starved ...
|
|
42909
|
Purification and characterization of pyrophosphate-dependent phosphofructokinase from phosphate-starved ...
|
|
42910
|
Purification and characterization of pyrophosphate-dependent phosphofructokinase from phosphate-starved ...
|
|
42911
|
Purification and characterization of pyrophosphate-dependent phosphofructokinase from phosphate-starved ...
|
|
42912
|
Purification and characterization of pyrophosphate-dependent phosphofructokinase from phosphate-starved ...
|
|
42913
|
Purification and characterization of pyrophosphate-dependent phosphofructokinase from phosphate-starved ...
|
|
42914
|
Purification and characterization of pyrophosphate-dependent phosphofructokinase from phosphate-starved ...
|
|
42915
|
Purification and characterization of pyrophosphate-dependent phosphofructokinase from phosphate-starved ...
|
|
42916
|
Purification and characterization of pyrophosphate-dependent phosphofructokinase from phosphate-starved ...
|
|
42917
|
Purification and characterization of pyrophosphate-dependent phosphofructokinase from phosphate-starved ...
|
|
42918
|
Purification and characterization of pyrophosphate-dependent phosphofructokinase from phosphate-starved ...
|
|
42919
|
Purification and characterization of pyrophosphate-dependent phosphofructokinase from phosphate-starved ...
|
|
42920
|
Purification and characterization of pyrophosphate-dependent phosphofructokinase from phosphate-starved ...
|
|
42921
|
Purification and characterization of pyrophosphate-dependent phosphofructokinase from phosphate-starved ...
|
|
42922
|
Purification and characterization of pyrophosphate-dependent phosphofructokinase from phosphate-starved ...
|
|
42923
|
Purification and characterization of pyrophosphate-dependent phosphofructokinase from phosphate-starved ...
|
|
42924
|
Purification and characterization of pyrophosphate-dependent phosphofructokinase from phosphate-starved ...
|
|
42925
|
Purification and characterization of pyrophosphate-dependent phosphofructokinase from phosphate-starved ...
|
|
42926
|
Purification and characterization of pyrophosphate-dependent phosphofructokinase from phosphate-starved ...
|
|
42927
|
Purification and characterization of pyrophosphate-dependent phosphofructokinase from phosphate-starved ...
|
|
42928
|
Purification and characterization of pyrophosphate-dependent phosphofructokinase from phosphate-starved ...
|
|
42929
|
Arabidopsis thaliana beta-Glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides
|
|
42930
|
Arabidopsis thaliana beta-Glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides
|
|
42931
|
Arabidopsis thaliana beta-Glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides
|
|
42932
|
Arabidopsis thaliana beta-Glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides
|
|
42933
|
Arabidopsis thaliana beta-Glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides
|
|
42934
|
Arabidopsis thaliana beta-Glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides
|
|
42935
|
Arabidopsis thaliana beta-Glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides
|
|
42936
|
Arabidopsis thaliana beta-Glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides
|
|
42937
|
Arabidopsis thaliana beta-Glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides
|
|
42938
|
Arabidopsis thaliana beta-Glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides
|
|
42939
|
Arabidopsis thaliana beta-Glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides
|
|
42940
|
Arabidopsis thaliana beta-Glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides
|
|
42941
|
Arabidopsis thaliana beta-Glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides
|
|
42942
|
Arabidopsis thaliana beta-Glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides
|
|
42943
|
Arabidopsis thaliana beta-Glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides
|
|
42944
|
Arabidopsis thaliana beta-Glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides
|
|
42945
|
Arabidopsis thaliana beta-Glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides
|
|
42946
|
Arabidopsis thaliana beta-Glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides
|
|
42947
|
Arabidopsis thaliana beta-Glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides
|
|
42948
|
Human arylacetamide deacetylase is a principal enzyme in flutamide hydrolysis
|
|
42949
|
Human arylacetamide deacetylase is a principal enzyme in flutamide hydrolysis
|
|
42950
|
Human arylacetamide deacetylase is a principal enzyme in flutamide hydrolysis
|
|
42951
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42952
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42953
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42954
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42955
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42956
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42957
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42958
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42959
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42960
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42961
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42962
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42963
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42964
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42965
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42966
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42967
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42968
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42969
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42970
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42971
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42972
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42973
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42974
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42975
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42976
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42977
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42978
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42979
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42980
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42981
|
An unusual peptide deformylase features in the human mitochondrial N-terminal methionine excision pathway
|
|
42982
|
Purification and characterization of hyoscyamine 6 beta-hydroxylase from root cultures of Hyoscyamus niger L. ...
|
|
42983
|
Purification and characterization of hyoscyamine 6 beta-hydroxylase from root cultures of Hyoscyamus niger L. ...
|
|
42984
|
Purification and characterization of hyoscyamine 6 beta-hydroxylase from root cultures of Hyoscyamus niger L. ...
|
|
42985
|
Purification and characterization of hyoscyamine 6 beta-hydroxylase from root cultures of Hyoscyamus niger L. ...
|
|
42986
|
Enzymes of phosphatidylcholine synthesis in lemna, soybean, and carrot
|
|
42987
|
Crystal structure of NADP(H)-dependent 1,5-anhydro-D-fructose reductase from Sinorhizobium morelense at 2.2 A ...
|
|
42988
|
Crystal structure of NADP(H)-dependent 1,5-anhydro-D-fructose reductase from Sinorhizobium morelense at 2.2 A ...
|
|
42989
|
Crystal structure of NADP(H)-dependent 1,5-anhydro-D-fructose reductase from Sinorhizobium morelense at 2.2 A ...
|
|
42990
|
Crystal structure of NADP(H)-dependent 1,5-anhydro-D-fructose reductase from Sinorhizobium morelense at 2.2 A ...
|
|
42991
|
Crystal structure of NADP(H)-dependent 1,5-anhydro-D-fructose reductase from Sinorhizobium morelense at 2.2 A ...
|
|
42992
|
Crystal structure of NADP(H)-dependent 1,5-anhydro-D-fructose reductase from Sinorhizobium morelense at 2.2 A ...
|
|
42993
|
Crystal structure of NADP(H)-dependent 1,5-anhydro-D-fructose reductase from Sinorhizobium morelense at 2.2 A ...
|
|
42994
|
Crystal structure of NADP(H)-dependent 1,5-anhydro-D-fructose reductase from Sinorhizobium morelense at 2.2 A ...
|
|
42995
|
Crystal structure of NADP(H)-dependent 1,5-anhydro-D-fructose reductase from Sinorhizobium morelense at 2.2 A ...
|
|
42996
|
Crystal structure of NADP(H)-dependent 1,5-anhydro-D-fructose reductase from Sinorhizobium morelense at 2.2 A ...
|
|
42997
|
Crystal structure of NADP(H)-dependent 1,5-anhydro-D-fructose reductase from Sinorhizobium morelense at 2.2 A ...
|
|
42998
|
Crystal structure of NADP(H)-dependent 1,5-anhydro-D-fructose reductase from Sinorhizobium morelense at 2.2 A ...
|
|
42999
|
Crystal structure of NADP(H)-dependent 1,5-anhydro-D-fructose reductase from Sinorhizobium morelense at 2.2 A ...
|
|
43000
|
Crystal structure of NADP(H)-dependent 1,5-anhydro-D-fructose reductase from Sinorhizobium morelense at 2.2 A ...
|





 If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.
If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.