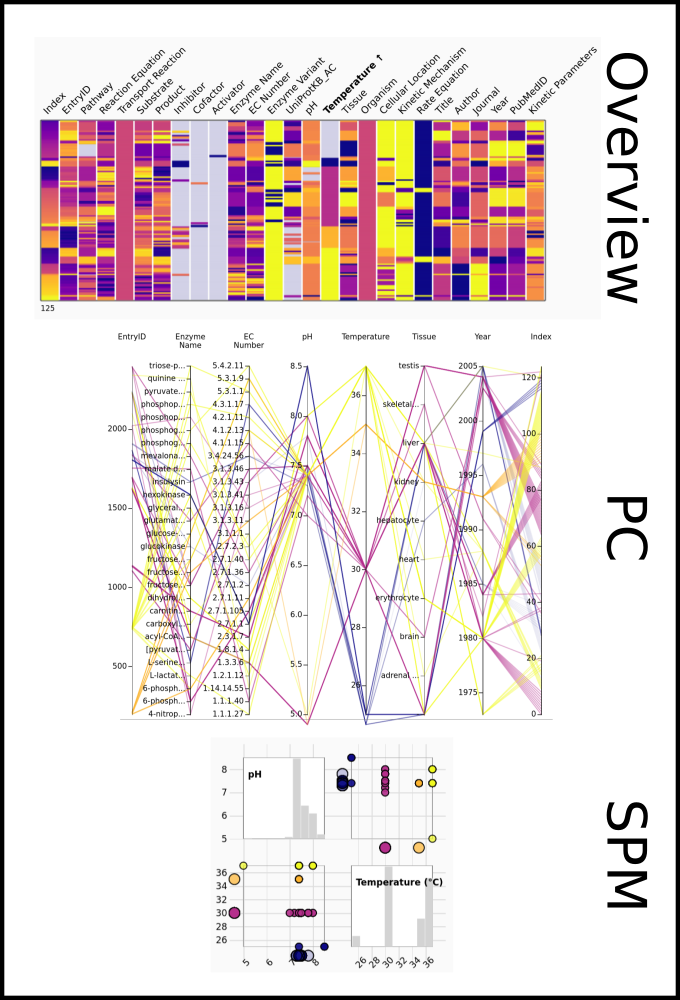
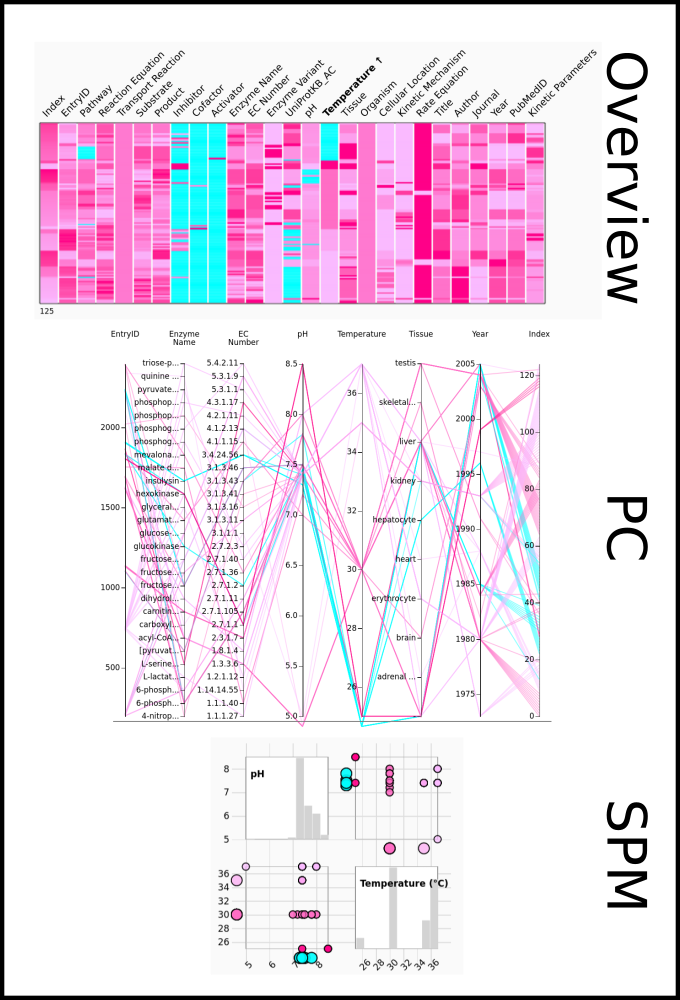
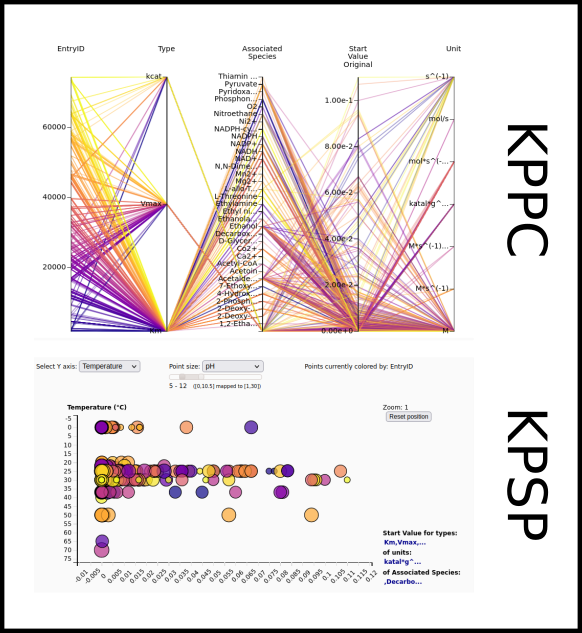
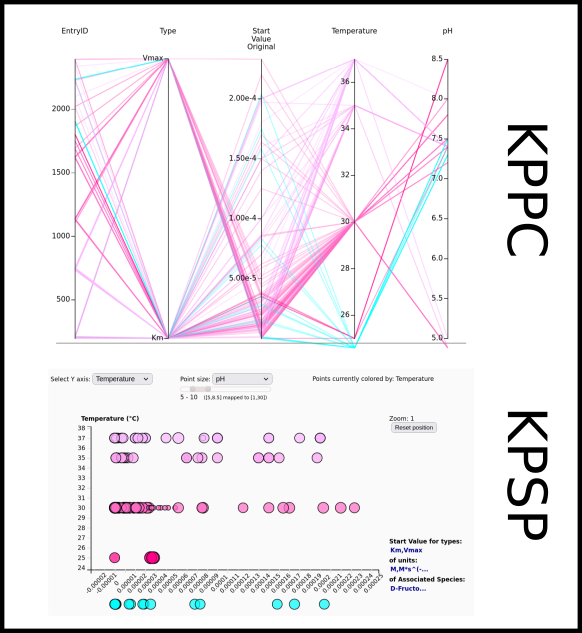
To improve SABIO-RK data interoperability, semantic markup was added to web pages as described and defined by
This structured information makes it easier to discover, collate and analyse our data.
|
37001
|
Purified gamma-glutamyl transpeptidases from tomato exhibit high affinity for glutathione and glutathione ...
|
|
37002
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37003
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37004
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37005
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37006
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37007
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37008
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37009
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37010
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37011
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37012
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37013
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37014
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37015
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37016
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37017
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37018
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37019
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37020
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37021
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37022
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37023
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37024
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37025
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37026
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37027
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37028
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37029
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37030
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37031
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37032
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37033
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37034
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37035
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37036
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37037
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37038
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37039
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37040
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37041
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37042
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37043
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37044
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37045
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37046
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37047
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37048
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37049
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37050
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37051
|
Correlations of inhibitor kinetics for Pneumocystis jirovecii and human dihydrofolate reductase with ...
|
|
37052
|
Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase ...
|
|
37053
|
Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase ...
|
|
37054
|
Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase ...
|
|
37055
|
Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase ...
|
|
37056
|
Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase ...
|
|
37057
|
Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase ...
|
|
37058
|
Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase ...
|
|
37059
|
Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase ...
|
|
37060
|
Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase ...
|
|
37061
|
Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase ...
|
|
37062
|
Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase ...
|
|
37063
|
A GTP-dependent vertebrate-type phosphoenolpyruvate carboxykinase from Mycobacterium smegmatis
|
|
37064
|
A GTP-dependent vertebrate-type phosphoenolpyruvate carboxykinase from Mycobacterium smegmatis
|
|
37065
|
A GTP-dependent vertebrate-type phosphoenolpyruvate carboxykinase from Mycobacterium smegmatis
|
|
37066
|
A GTP-dependent vertebrate-type phosphoenolpyruvate carboxykinase from Mycobacterium smegmatis
|
|
37067
|
A GTP-dependent vertebrate-type phosphoenolpyruvate carboxykinase from Mycobacterium smegmatis
|
|
37068
|
A GTP-dependent vertebrate-type phosphoenolpyruvate carboxykinase from Mycobacterium smegmatis
|
|
37069
|
A GTP-dependent vertebrate-type phosphoenolpyruvate carboxykinase from Mycobacterium smegmatis
|
|
37070
|
A GTP-dependent vertebrate-type phosphoenolpyruvate carboxykinase from Mycobacterium smegmatis
|
|
37071
|
A GTP-dependent vertebrate-type phosphoenolpyruvate carboxykinase from Mycobacterium smegmatis
|
|
37072
|
A GTP-dependent vertebrate-type phosphoenolpyruvate carboxykinase from Mycobacterium smegmatis
|
|
37073
|
A GTP-dependent vertebrate-type phosphoenolpyruvate carboxykinase from Mycobacterium smegmatis
|
|
37074
|
A GTP-dependent vertebrate-type phosphoenolpyruvate carboxykinase from Mycobacterium smegmatis
|
|
37075
|
A GTP-dependent vertebrate-type phosphoenolpyruvate carboxykinase from Mycobacterium smegmatis
|
|
37076
|
A GTP-dependent vertebrate-type phosphoenolpyruvate carboxykinase from Mycobacterium smegmatis
|
|
37077
|
A GTP-dependent vertebrate-type phosphoenolpyruvate carboxykinase from Mycobacterium smegmatis
|
|
37078
|
A GTP-dependent vertebrate-type phosphoenolpyruvate carboxykinase from Mycobacterium smegmatis
|
|
37079
|
A GTP-dependent vertebrate-type phosphoenolpyruvate carboxykinase from Mycobacterium smegmatis
|
|
37080
|
A GTP-dependent vertebrate-type phosphoenolpyruvate carboxykinase from Mycobacterium smegmatis
|
|
37081
|
A GTP-dependent vertebrate-type phosphoenolpyruvate carboxykinase from Mycobacterium smegmatis
|
|
37082
|
A GTP-dependent vertebrate-type phosphoenolpyruvate carboxykinase from Mycobacterium smegmatis
|
|
37083
|
Purification and characterization of neutral and alkaline invertase from carrot
|
|
37084
|
Purification and characterization of neutral and alkaline invertase from carrot
|
|
37085
|
Purification and characterization of neutral and alkaline invertase from carrot
|
|
37086
|
Purification and characterization of neutral and alkaline invertase from carrot
|
|
37087
|
Purification and characterization of neutral and alkaline invertase from carrot
|
|
37088
|
Purification and characterization of neutral and alkaline invertase from carrot
|
|
37089
|
Purification and characterization of neutral and alkaline invertase from carrot
|
|
37090
|
Purification and characterization of neutral and alkaline invertase from carrot
|
|
37091
|
Purification and characterization of neutral and alkaline invertase from carrot
|
|
37092
|
Purification and characterization of neutral and alkaline invertase from carrot
|
|
37093
|
Purification and characterization of neutral and alkaline invertase from carrot
|
|
37094
|
Purification and characterization of neutral and alkaline invertase from carrot
|
|
37095
|
Purification and characterization of neutral and alkaline invertase from carrot
|
|
37096
|
Purification and characterization of neutral and alkaline invertase from carrot
|
|
37097
|
Purification and characterization of neutral and alkaline invertase from carrot
|
|
37098
|
Purification and characterization of neutral and alkaline invertase from carrot
|
|
37099
|
Purification and characterization of neutral and alkaline invertase from carrot
|
|
37100
|
Purification and characterization of neutral and alkaline invertase from carrot
|
|
37101
|
Purification and characterization of neutral and alkaline invertase from carrot
|
|
37102
|
Purification and characterization of neutral and alkaline invertase from carrot
|
|
37103
|
Purification and characterization of neutral and alkaline invertase from carrot
|
|
37104
|
Purification and characterization of neutral and alkaline invertase from carrot
|
|
37105
|
Complete reconstitution of the human coenzyme A biosynthetic pathway via comparative genomics
|
|
37106
|
Complete reconstitution of the human coenzyme A biosynthetic pathway via comparative genomics
|
|
37107
|
Complete reconstitution of the human coenzyme A biosynthetic pathway via comparative genomics
|
|
37108
|
Complete reconstitution of the human coenzyme A biosynthetic pathway via comparative genomics
|
|
37109
|
Complete reconstitution of the human coenzyme A biosynthetic pathway via comparative genomics
|
|
37110
|
Complete reconstitution of the human coenzyme A biosynthetic pathway via comparative genomics
|
|
37111
|
Involvement of glutamate 399 and lysine 192 in the mechanism of human liver mitochondrial aldehyde ...
|
|
37112
|
Involvement of glutamate 399 and lysine 192 in the mechanism of human liver mitochondrial aldehyde ...
|
|
37113
|
Involvement of glutamate 399 and lysine 192 in the mechanism of human liver mitochondrial aldehyde ...
|
|
37114
|
Involvement of glutamate 399 and lysine 192 in the mechanism of human liver mitochondrial aldehyde ...
|
|
37115
|
Involvement of glutamate 399 and lysine 192 in the mechanism of human liver mitochondrial aldehyde ...
|
|
37116
|
Involvement of glutamate 399 and lysine 192 in the mechanism of human liver mitochondrial aldehyde ...
|
|
37117
|
Involvement of glutamate 399 and lysine 192 in the mechanism of human liver mitochondrial aldehyde ...
|
|
37118
|
Involvement of glutamate 399 and lysine 192 in the mechanism of human liver mitochondrial aldehyde ...
|
|
37119
|
Involvement of glutamate 399 and lysine 192 in the mechanism of human liver mitochondrial aldehyde ...
|
|
37120
|
Involvement of glutamate 399 and lysine 192 in the mechanism of human liver mitochondrial aldehyde ...
|
|
37121
|
Involvement of glutamate 399 and lysine 192 in the mechanism of human liver mitochondrial aldehyde ...
|
|
37122
|
Involvement of glutamate 399 and lysine 192 in the mechanism of human liver mitochondrial aldehyde ...
|
|
37123
|
Involvement of glutamate 399 and lysine 192 in the mechanism of human liver mitochondrial aldehyde ...
|
|
37124
|
Involvement of glutamate 399 and lysine 192 in the mechanism of human liver mitochondrial aldehyde ...
|
|
37125
|
Ether lipid synthesis: purification and identification of alkyl dihydroxyacetone phosphate synthase from ...
|
|
37126
|
Ether lipid synthesis: purification and identification of alkyl dihydroxyacetone phosphate synthase from ...
|
|
37127
|
Ether lipid synthesis: purification and identification of alkyl dihydroxyacetone phosphate synthase from ...
|
|
37128
|
Ether lipid synthesis: purification and identification of alkyl dihydroxyacetone phosphate synthase from ...
|
|
37129
|
Ether lipid synthesis: purification and identification of alkyl dihydroxyacetone phosphate synthase from ...
|
|
37130
|
Genetic and biochemical characterization of the chromosome-encoded class B beta-lactamases from Shewanella ...
|
|
37131
|
Genetic and biochemical characterization of the chromosome-encoded class B beta-lactamases from Shewanella ...
|
|
37132
|
Genetic and biochemical characterization of the chromosome-encoded class B beta-lactamases from Shewanella ...
|
|
37133
|
Genetic and biochemical characterization of the chromosome-encoded class B beta-lactamases from Shewanella ...
|
|
37134
|
Genetic and biochemical characterization of the chromosome-encoded class B beta-lactamases from Shewanella ...
|
|
37135
|
Genetic and biochemical characterization of the chromosome-encoded class B beta-lactamases from Shewanella ...
|
|
37136
|
Genetic and biochemical characterization of the chromosome-encoded class B beta-lactamases from Shewanella ...
|
|
37137
|
Genetic and biochemical characterization of the chromosome-encoded class B beta-lactamases from Shewanella ...
|
|
37138
|
Genetic and biochemical characterization of the chromosome-encoded class B beta-lactamases from Shewanella ...
|
|
37139
|
Genetic and biochemical characterization of the chromosome-encoded class B beta-lactamases from Shewanella ...
|
|
37140
|
Genetic and biochemical characterization of the chromosome-encoded class B beta-lactamases from Shewanella ...
|
|
37141
|
Genetic and biochemical characterization of the chromosome-encoded class B beta-lactamases from Shewanella ...
|
|
37142
|
Genetic and biochemical characterization of the chromosome-encoded class B beta-lactamases from Shewanella ...
|
|
37143
|
Genetic and biochemical characterization of the chromosome-encoded class B beta-lactamases from Shewanella ...
|
|
37144
|
Genetic and biochemical characterization of the chromosome-encoded class B beta-lactamases from Shewanella ...
|
|
37145
|
Genetic and biochemical characterization of the chromosome-encoded class B beta-lactamases from Shewanella ...
|
|
37146
|
Genetic and biochemical characterization of the chromosome-encoded class B beta-lactamases from Shewanella ...
|
|
37147
|
Genetic and biochemical characterization of the chromosome-encoded class B beta-lactamases from Shewanella ...
|
|
37148
|
Genetic and biochemical characterization of the chromosome-encoded class B beta-lactamases from Shewanella ...
|
|
37149
|
Genetic and biochemical characterization of the chromosome-encoded class B beta-lactamases from Shewanella ...
|
|
37150
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37151
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37152
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37153
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37154
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37155
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37156
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37157
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37158
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37159
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37160
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37161
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37162
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37163
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37164
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37165
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37166
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37167
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37168
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37169
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37170
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37171
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37172
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37173
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37174
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37175
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37176
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37177
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37178
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37179
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37180
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37181
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37182
|
Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and ...
|
|
37183
|
Genetic and biochemical characterization of a chromosome-encoded carbapenem-hydrolyzing ambler class D ...
|
|
37184
|
Genetic and biochemical characterization of a chromosome-encoded carbapenem-hydrolyzing ambler class D ...
|
|
37185
|
Genetic and biochemical characterization of a chromosome-encoded carbapenem-hydrolyzing ambler class D ...
|
|
37186
|
Genetic and biochemical characterization of a chromosome-encoded carbapenem-hydrolyzing ambler class D ...
|
|
37187
|
Genetic and biochemical characterization of a chromosome-encoded carbapenem-hydrolyzing ambler class D ...
|
|
37188
|
Genetic and biochemical characterization of a chromosome-encoded carbapenem-hydrolyzing ambler class D ...
|
|
37189
|
Genetic and biochemical characterization of a chromosome-encoded carbapenem-hydrolyzing ambler class D ...
|
|
37190
|
Genetic and biochemical characterization of a chromosome-encoded carbapenem-hydrolyzing ambler class D ...
|
|
37191
|
Genetic and biochemical characterization of a chromosome-encoded carbapenem-hydrolyzing ambler class D ...
|
|
37192
|
Genetic and biochemical characterization of a chromosome-encoded carbapenem-hydrolyzing ambler class D ...
|
|
37193
|
Genetic and biochemical characterization of a chromosome-encoded carbapenem-hydrolyzing ambler class D ...
|
|
37194
|
Genetic and biochemical characterization of a chromosome-encoded carbapenem-hydrolyzing ambler class D ...
|
|
37195
|
Genetic and biochemical characterization of a chromosome-encoded carbapenem-hydrolyzing ambler class D ...
|
|
37196
|
Genetic and biochemical characterization of a chromosome-encoded carbapenem-hydrolyzing ambler class D ...
|
|
37197
|
Genetic and biochemical characterization of a chromosome-encoded carbapenem-hydrolyzing ambler class D ...
|
|
37198
|
Genetic and biochemical characterization of a chromosome-encoded carbapenem-hydrolyzing ambler class D ...
|
|
37199
|
A switch in the kinase domain of rat testis 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase
|
|
37200
|
A switch in the kinase domain of rat testis 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase
|
|
37201
|
A switch in the kinase domain of rat testis 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase
|
|
37202
|
A switch in the kinase domain of rat testis 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase
|
|
37203
|
A switch in the kinase domain of rat testis 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase
|
|
37204
|
A switch in the kinase domain of rat testis 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase
|
|
37205
|
A switch in the kinase domain of rat testis 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase
|
|
37206
|
A switch in the kinase domain of rat testis 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase
|
|
37207
|
A switch in the kinase domain of rat testis 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase
|
|
37208
|
A switch in the kinase domain of rat testis 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase
|
|
37209
|
A switch in the kinase domain of rat testis 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase
|
|
37210
|
Isolation and characterization of human liver aldehyde dehydrogenase
|
|
37211
|
Isolation and characterization of human liver aldehyde dehydrogenase
|
|
37212
|
Isolation and characterization of human liver aldehyde dehydrogenase
|
|
37213
|
Isolation and characterization of human liver aldehyde dehydrogenase
|
|
37214
|
Isolation and characterization of human liver aldehyde dehydrogenase
|
|
37215
|
Isolation and characterization of human liver aldehyde dehydrogenase
|
|
37216
|
Isolation and characterization of human liver aldehyde dehydrogenase
|
|
37217
|
Isolation and characterization of human liver aldehyde dehydrogenase
|
|
37218
|
Isolation and characterization of human liver aldehyde dehydrogenase
|
|
37219
|
Isolation and characterization of human liver aldehyde dehydrogenase
|
|
37220
|
Isolation and characterization of human liver aldehyde dehydrogenase
|
|
37221
|
Isolation and characterization of human liver aldehyde dehydrogenase
|
|
37222
|
Isolation and characterization of human liver aldehyde dehydrogenase
|
|
37223
|
Isolation and characterization of human liver aldehyde dehydrogenase
|
|
37224
|
Isolation and characterization of human liver aldehyde dehydrogenase
|
|
37225
|
Isolation and characterization of human liver aldehyde dehydrogenase
|
|
37226
|
Isolation and characterization of human liver aldehyde dehydrogenase
|
|
37227
|
Isolation and characterization of human liver aldehyde dehydrogenase
|
|
37228
|
Isolation and characterization of human liver aldehyde dehydrogenase
|
|
37229
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37230
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37231
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37232
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37233
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37234
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37235
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37236
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37237
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37238
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37239
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37240
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37241
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37242
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37243
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37244
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37245
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37246
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37247
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37248
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37249
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37250
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37251
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37252
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37253
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37254
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37255
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37256
|
Beta-ketophosphonates as beta-lactamase inhibitors: Intramolecular cooperativity between the hydrophobic ...
|
|
37257
|
Purification and characterization of the alcohol dehydrogenase with a broad substrate specificity originated ...
|
|
37258
|
Purification and characterization of the alcohol dehydrogenase with a broad substrate specificity originated ...
|
|
37259
|
Purification and characterization of the alcohol dehydrogenase with a broad substrate specificity originated ...
|
|
37260
|
Purification and characterization of the alcohol dehydrogenase with a broad substrate specificity originated ...
|
|
37261
|
Purification and characterization of the alcohol dehydrogenase with a broad substrate specificity originated ...
|
|
37262
|
Purification and characterization of the alcohol dehydrogenase with a broad substrate specificity originated ...
|
|
37263
|
Purification and characterization of the alcohol dehydrogenase with a broad substrate specificity originated ...
|
|
37264
|
Purification and characterization of the alcohol dehydrogenase with a broad substrate specificity originated ...
|
|
37265
|
Purification and characterization of the alcohol dehydrogenase with a broad substrate specificity originated ...
|
|
37266
|
Purification and substrate specificity of honeybee, Apis mellifera L., alpha-glucosidase III
|
|
37267
|
Purification and substrate specificity of honeybee, Apis mellifera L., alpha-glucosidase III
|
|
37268
|
Purification and substrate specificity of honeybee, Apis mellifera L., alpha-glucosidase III
|
|
37269
|
Purification and substrate specificity of honeybee, Apis mellifera L., alpha-glucosidase III
|
|
37270
|
Purification and substrate specificity of honeybee, Apis mellifera L., alpha-glucosidase III
|
|
37271
|
Purification and substrate specificity of honeybee, Apis mellifera L., alpha-glucosidase III
|
|
37272
|
Purification and substrate specificity of honeybee, Apis mellifera L., alpha-glucosidase III
|
|
37273
|
Purification and substrate specificity of honeybee, Apis mellifera L., alpha-glucosidase III
|
|
37274
|
Purification and substrate specificity of honeybee, Apis mellifera L., alpha-glucosidase III
|
|
37275
|
Purification and substrate specificity of honeybee, Apis mellifera L., alpha-glucosidase III
|
|
37276
|
Purification and substrate specificity of honeybee, Apis mellifera L., alpha-glucosidase III
|
|
37277
|
Purification and substrate specificity of honeybee, Apis mellifera L., alpha-glucosidase III
|
|
37278
|
Purification and substrate specificity of honeybee, Apis mellifera L., alpha-glucosidase III
|
|
37279
|
Multitasking in signal transduction by a promiscuous human Ins(3,4,5,6)P(4) 1-kinase/Ins(1,3,4)P(3) 5/6-kinase
|
|
37280
|
Multitasking in signal transduction by a promiscuous human Ins(3,4,5,6)P(4) 1-kinase/Ins(1,3,4)P(3) 5/6-kinase
|
|
37281
|
A single cyclohexadienyl dehydratase specifies the prephenate dehydratase and arogenate dehydratase components ...
|
|
37282
|
A single cyclohexadienyl dehydratase specifies the prephenate dehydratase and arogenate dehydratase components ...
|
|
37283
|
The mechanism of ATP inhibition of wild type and mutant phosphofructo-1-kinase from Escherichia coli
|
|
37284
|
The mechanism of ATP inhibition of wild type and mutant phosphofructo-1-kinase from Escherichia coli
|
|
37285
|
The mechanism of ATP inhibition of wild type and mutant phosphofructo-1-kinase from Escherichia coli
|
|
37286
|
The mechanism of ATP inhibition of wild type and mutant phosphofructo-1-kinase from Escherichia coli
|
|
37287
|
The mechanism of ATP inhibition of wild type and mutant phosphofructo-1-kinase from Escherichia coli
|
|
37288
|
The mechanism of ATP inhibition of wild type and mutant phosphofructo-1-kinase from Escherichia coli
|
|
37289
|
The mechanism of ATP inhibition of wild type and mutant phosphofructo-1-kinase from Escherichia coli
|
|
37290
|
The mechanism of ATP inhibition of wild type and mutant phosphofructo-1-kinase from Escherichia coli
|
|
37291
|
The mechanism of ATP inhibition of wild type and mutant phosphofructo-1-kinase from Escherichia coli
|
|
37292
|
The mechanism of ATP inhibition of wild type and mutant phosphofructo-1-kinase from Escherichia coli
|
|
37293
|
The mechanism of ATP inhibition of wild type and mutant phosphofructo-1-kinase from Escherichia coli
|
|
37294
|
The mechanism of ATP inhibition of wild type and mutant phosphofructo-1-kinase from Escherichia coli
|
|
37295
|
The mechanism of ATP inhibition of wild type and mutant phosphofructo-1-kinase from Escherichia coli
|
|
37296
|
Kinetic and structural properties of triosephosphate isomerase from Helicobacter pylori
|
|
37297
|
Kinetic and structural properties of triosephosphate isomerase from Helicobacter pylori
|
|
37298
|
Kinetic and structural properties of triosephosphate isomerase from Helicobacter pylori
|
|
37299
|
Kinetic and structural properties of triosephosphate isomerase from Helicobacter pylori
|
|
37300
|
Kinetic and structural properties of triosephosphate isomerase from Helicobacter pylori
|
|
37301
|
Sequence, expression, and characterization of the first archaeal ATP-dependent 6-phosphofructokinase, a ...
|
|
37302
|
Sequence, expression, and characterization of the first archaeal ATP-dependent 6-phosphofructokinase, a ...
|
|
37303
|
Sequence, expression, and characterization of the first archaeal ATP-dependent 6-phosphofructokinase, a ...
|
|
37304
|
Sequence, expression, and characterization of the first archaeal ATP-dependent 6-phosphofructokinase, a ...
|
|
37305
|
Sequence, expression, and characterization of the first archaeal ATP-dependent 6-phosphofructokinase, a ...
|
|
37306
|
Sequence, expression, and characterization of the first archaeal ATP-dependent 6-phosphofructokinase, a ...
|
|
37307
|
Sequence, expression, and characterization of the first archaeal ATP-dependent 6-phosphofructokinase, a ...
|
|
37308
|
Sequence, expression, and characterization of the first archaeal ATP-dependent 6-phosphofructokinase, a ...
|
|
37309
|
Sequence, expression, and characterization of the first archaeal ATP-dependent 6-phosphofructokinase, a ...
|
|
37310
|
Sequence, expression, and characterization of the first archaeal ATP-dependent 6-phosphofructokinase, a ...
|
|
37311
|
Pig red blood cell hexokinase: regulatory characteristics and possible physiological role
|
|
37312
|
Pig red blood cell hexokinase: regulatory characteristics and possible physiological role
|
|
37313
|
Pig red blood cell hexokinase: regulatory characteristics and possible physiological role
|
|
37314
|
Pig red blood cell hexokinase: regulatory characteristics and possible physiological role
|
|
37315
|
Pig red blood cell hexokinase: regulatory characteristics and possible physiological role
|
|
37316
|
Pig red blood cell hexokinase: regulatory characteristics and possible physiological role
|
|
37317
|
Pig red blood cell hexokinase: regulatory characteristics and possible physiological role
|
|
37318
|
Pig red blood cell hexokinase: regulatory characteristics and possible physiological role
|
|
37319
|
Pig red blood cell hexokinase: regulatory characteristics and possible physiological role
|
|
37320
|
Pig red blood cell hexokinase: regulatory characteristics and possible physiological role
|
|
37321
|
Pig red blood cell hexokinase: regulatory characteristics and possible physiological role
|
|
37322
|
Pig red blood cell hexokinase: regulatory characteristics and possible physiological role
|
|
37323
|
Pig red blood cell hexokinase: regulatory characteristics and possible physiological role
|
|
37324
|
Pig red blood cell hexokinase: regulatory characteristics and possible physiological role
|
|
37325
|
Pig red blood cell hexokinase: regulatory characteristics and possible physiological role
|
|
37326
|
Evidence supporting a cis-enediol-based mechanism for Pyrococcus furiosus phosphoglucose isomerase
|
|
37327
|
Evidence supporting a cis-enediol-based mechanism for Pyrococcus furiosus phosphoglucose isomerase
|
|
37328
|
Evidence supporting a cis-enediol-based mechanism for Pyrococcus furiosus phosphoglucose isomerase
|
|
37329
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37330
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37331
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37332
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37333
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37334
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37335
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37336
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37337
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37338
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37339
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37340
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37341
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37342
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37343
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37344
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37345
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37346
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37347
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37348
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37349
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37350
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37351
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37352
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37353
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37354
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37355
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37356
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37357
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37358
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37359
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37360
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37361
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37362
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37363
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37364
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37365
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37366
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37367
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37368
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37369
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37370
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37371
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37372
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37373
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37374
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37375
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37376
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37377
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37378
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37379
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37380
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37381
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37382
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37383
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37384
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37385
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37386
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37387
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37388
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37389
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37390
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37391
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37392
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37393
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37394
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37395
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37396
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37397
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37398
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37399
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37400
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37401
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37402
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37403
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37404
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37405
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37406
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37407
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37408
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37409
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37410
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37411
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37412
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37413
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37414
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37415
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37416
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37417
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37418
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37419
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37420
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37421
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37422
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37423
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37424
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37425
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37426
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37427
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37428
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37429
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37430
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37431
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37432
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37433
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37434
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37435
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37436
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37437
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37438
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37439
|
Molecular cloning of a novel type of rat cytoplasmic 17beta-hydroxysteroid dehydrogenase distinct from the ...
|
|
37441
|
Purification and properties of the aspartyl ribonucleic acid synthetase of Lactobacillus arabinosus
|
|
37442
|
Purification and properties of the aspartyl ribonucleic acid synthetase of Lactobacillus arabinosus
|
|
37443
|
Purification and properties of the aspartyl ribonucleic acid synthetase of Lactobacillus arabinosus
|
|
37444
|
Pentose fermentation by Lactobacillus plantarum. II. L-arabinose isomerase
|
|
37445
|
Pentose fermentation by Lactobacillus plantarum. II. L-arabinose isomerase
|
|
37446
|
Characterization of a new extended-spectrum beta-lactamase (TEM-87) isolated in Proteus mirabilis during an ...
|
|
37447
|
Characterization of a new extended-spectrum beta-lactamase (TEM-87) isolated in Proteus mirabilis during an ...
|
|
37448
|
Characterization of a new extended-spectrum beta-lactamase (TEM-87) isolated in Proteus mirabilis during an ...
|
|
37449
|
Characterization of a new extended-spectrum beta-lactamase (TEM-87) isolated in Proteus mirabilis during an ...
|
|
37450
|
Characterization of a new extended-spectrum beta-lactamase (TEM-87) isolated in Proteus mirabilis during an ...
|
|
37451
|
Characterization of a new extended-spectrum beta-lactamase (TEM-87) isolated in Proteus mirabilis during an ...
|
|
37452
|
Characterization of a new extended-spectrum beta-lactamase (TEM-87) isolated in Proteus mirabilis during an ...
|
|
37453
|
Characterization of a new extended-spectrum beta-lactamase (TEM-87) isolated in Proteus mirabilis during an ...
|
|
37454
|
Characterization of a new extended-spectrum beta-lactamase (TEM-87) isolated in Proteus mirabilis during an ...
|
|
37455
|
Characterization of a new extended-spectrum beta-lactamase (TEM-87) isolated in Proteus mirabilis during an ...
|
|
37456
|
Characterization of a new extended-spectrum beta-lactamase (TEM-87) isolated in Proteus mirabilis during an ...
|
|
37457
|
Characterization of a new extended-spectrum beta-lactamase (TEM-87) isolated in Proteus mirabilis during an ...
|
|
37458
|
Activation of thiamin diphosphate and FAD in the phosphatedependent pyruvate oxidase from Lactobacillus ...
|
|
37459
|
Activation of thiamin diphosphate and FAD in the phosphatedependent pyruvate oxidase from Lactobacillus ...
|
|
37460
|
PPi-dependent phosphofructotransferase (phosphofructokinase) activity in the mollicutes (mycoplasma) ...
|
|
37461
|
Cloning, expression, and chromosomal localization of mouse liver bile acid CoA:amino acid N-acyltransferase
|
|
37463
|
The two acetyl-coenzyme A synthetases of Saccharomyces cerevisiae differ with respect to kinetic properties ...
|
|
37464
|
The two acetyl-coenzyme A synthetases of Saccharomyces cerevisiae differ with respect to kinetic properties ...
|
|
37465
|
The two acetyl-coenzyme A synthetases of Saccharomyces cerevisiae differ with respect to kinetic properties ...
|
|
37466
|
The two acetyl-coenzyme A synthetases of Saccharomyces cerevisiae differ with respect to kinetic properties ...
|
|
37467
|
The two acetyl-coenzyme A synthetases of Saccharomyces cerevisiae differ with respect to kinetic properties ...
|
|
37468
|
The two acetyl-coenzyme A synthetases of Saccharomyces cerevisiae differ with respect to kinetic properties ...
|
|
37469
|
Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase ...
|
|
37470
|
Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase ...
|
|
37471
|
Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase ...
|
|
37472
|
Purification and characterization of adenosine diphosphate ribose pyrophosphatase from human erythrocytes
|
|
37473
|
Purification and characterization of adenosine diphosphate ribose pyrophosphatase from human erythrocytes
|
|
37474
|
Expression, purification, and characterization of the human squalene synthase: use of yeast and baculoviral ...
|
|
37475
|
Expression, purification, and characterization of the human squalene synthase: use of yeast and baculoviral ...
|
|
37476
|
Expression, purification, and characterization of the human squalene synthase: use of yeast and baculoviral ...
|
|
37477
|
Expression, purification, and characterization of the human squalene synthase: use of yeast and baculoviral ...
|
|
37478
|
Expression, purification, and characterization of the human squalene synthase: use of yeast and baculoviral ...
|
|
37479
|
AtCHX13 is a plasma membrane K+ transporter
|
|
37480
|
AtCHX13 is a plasma membrane K+ transporter
|
|
37481
|
Ceftazidime and aztreonam resistance in Providencia stuartii: characterization of a natural TEM-derived ...
|
|
37482
|
Ceftazidime and aztreonam resistance in Providencia stuartii: characterization of a natural TEM-derived ...
|
|
37483
|
Ceftazidime and aztreonam resistance in Providencia stuartii: characterization of a natural TEM-derived ...
|
|
37484
|
Ceftazidime and aztreonam resistance in Providencia stuartii: characterization of a natural TEM-derived ...
|
|
37485
|
Ceftazidime and aztreonam resistance in Providencia stuartii: characterization of a natural TEM-derived ...
|
|
37486
|
Ceftazidime and aztreonam resistance in Providencia stuartii: characterization of a natural TEM-derived ...
|
|
37487
|
Ceftazidime and aztreonam resistance in Providencia stuartii: characterization of a natural TEM-derived ...
|
|
37488
|
Ceftazidime and aztreonam resistance in Providencia stuartii: characterization of a natural TEM-derived ...
|
|
37489
|
Ceftazidime and aztreonam resistance in Providencia stuartii: characterization of a natural TEM-derived ...
|
|
37490
|
Ceftazidime and aztreonam resistance in Providencia stuartii: characterization of a natural TEM-derived ...
|
|
37491
|
Ceftazidime and aztreonam resistance in Providencia stuartii: characterization of a natural TEM-derived ...
|
|
37492
|
Ceftazidime and aztreonam resistance in Providencia stuartii: characterization of a natural TEM-derived ...
|
|
37493
|
Ceftazidime and aztreonam resistance in Providencia stuartii: characterization of a natural TEM-derived ...
|
|
37494
|
Ceftazidime and aztreonam resistance in Providencia stuartii: characterization of a natural TEM-derived ...
|
|
37495
|
Ceftazidime and aztreonam resistance in Providencia stuartii: characterization of a natural TEM-derived ...
|
|
37496
|
Ceftazidime and aztreonam resistance in Providencia stuartii: characterization of a natural TEM-derived ...
|
|
37497
|
Ceftazidime and aztreonam resistance in Providencia stuartii: characterization of a natural TEM-derived ...
|
|
37498
|
Ceftazidime and aztreonam resistance in Providencia stuartii: characterization of a natural TEM-derived ...
|
|
37499
|
Purification and characterization of cholyl-CoA: taurine N-acetyltransferase from the liver of domestic fowl ...
|
|
37500
|
Purification and characterization of cholyl-CoA: taurine N-acetyltransferase from the liver of domestic fowl ...
|
|
37501
|
Human and porcine aminoacylase I overproduced in a baculovirus expression vector system: evidence for ...
|
|
37502
|
Human and porcine aminoacylase I overproduced in a baculovirus expression vector system: evidence for ...
|
|
37503
|
Human and porcine aminoacylase I overproduced in a baculovirus expression vector system: evidence for ...
|
|
37504
|
Human and porcine aminoacylase I overproduced in a baculovirus expression vector system: evidence for ...
|
|
37506
|
Purification and characterization of a microsomal bile acid beta-glucosidase from human liver
|
|
37507
|
Purification and characterization of a microsomal bile acid beta-glucosidase from human liver
|
|
37508
|
Purification and characterization of a microsomal bile acid beta-glucosidase from human liver
|
|
37509
|
Purification and characterization of a microsomal bile acid beta-glucosidase from human liver
|
|
37510
|
Purification and characterization of a microsomal bile acid beta-glucosidase from human liver
|
|
37511
|
Purification and characterization of a microsomal bile acid beta-glucosidase from human liver
|
|
37512
|
Purification and characterization of a microsomal bile acid beta-glucosidase from human liver
|
|
37513
|
Purification and characterization of a microsomal bile acid beta-glucosidase from human liver
|
|
37514
|
Purification and characterization of a microsomal bile acid beta-glucosidase from human liver
|
|
37515
|
Purification and characterization of a microsomal bile acid beta-glucosidase from human liver
|
|
37516
|
Kinetics of activation of L-lactate dehydrogenase from Streptococcus faecalis by fructose 1,6-bisphosphate and ...
|
|
37517
|
Kinetics of activation of L-lactate dehydrogenase from Streptococcus faecalis by fructose 1,6-bisphosphate and ...
|
|
37518
|
Kinetics of activation of L-lactate dehydrogenase from Streptococcus faecalis by fructose 1,6-bisphosphate and ...
|
|
37519
|
Kinetics of activation of L-lactate dehydrogenase from Streptococcus faecalis by fructose 1,6-bisphosphate and ...
|
|
37520
|
Kinetics of activation of L-lactate dehydrogenase from Streptococcus faecalis by fructose 1,6-bisphosphate and ...
|
|
37521
|
Kinetics of activation of L-lactate dehydrogenase from Streptococcus faecalis by fructose 1,6-bisphosphate and ...
|
|
37522
|
Kinetics of activation of L-lactate dehydrogenase from Streptococcus faecalis by fructose 1,6-bisphosphate and ...
|
|
37523
|
Kinetics of activation of L-lactate dehydrogenase from Streptococcus faecalis by fructose 1,6-bisphosphate and ...
|
|
37524
|
Kinetics of activation of L-lactate dehydrogenase from Streptococcus faecalis by fructose 1,6-bisphosphate and ...
|
|
37525
|
The Ins(1,3,4)P3 5/6-kinase/Ins(3,4,5,6)P4 1-kinase is not a protein kinase
|
|
37526
|
The Ins(1,3,4)P3 5/6-kinase/Ins(3,4,5,6)P4 1-kinase is not a protein kinase
|
|
37527
|
The Ins(1,3,4)P3 5/6-kinase/Ins(3,4,5,6)P4 1-kinase is not a protein kinase
|
|
37528
|
The Ins(1,3,4)P3 5/6-kinase/Ins(3,4,5,6)P4 1-kinase is not a protein kinase
|
|
37529
|
The Ins(1,3,4)P3 5/6-kinase/Ins(3,4,5,6)P4 1-kinase is not a protein kinase
|
|
37530
|
The Ins(1,3,4)P3 5/6-kinase/Ins(3,4,5,6)P4 1-kinase is not a protein kinase
|
|
37531
|
The Ins(1,3,4)P3 5/6-kinase/Ins(3,4,5,6)P4 1-kinase is not a protein kinase
|
|
37532
|
The Ins(1,3,4)P3 5/6-kinase/Ins(3,4,5,6)P4 1-kinase is not a protein kinase
|
|
37533
|
The Ins(1,3,4)P3 5/6-kinase/Ins(3,4,5,6)P4 1-kinase is not a protein kinase
|
|
37534
|
Peroxisome proliferator-induced acyl-CoA thioesterase from rat liver cytosol: molecular cloning and functional ...
|
|
37535
|
Peroxisome proliferator-induced acyl-CoA thioesterase from rat liver cytosol: molecular cloning and functional ...
|
|
37536
|
Purification and characterization of bile acid-CoA:amino acid N-acyltransferase from human liver
|
|
37537
|
Purification and characterization of bile acid-CoA:amino acid N-acyltransferase from human liver
|
|
37538
|
Purification and characterization of bile acid-CoA:amino acid N-acyltransferase from human liver
|
|
37539
|
Purification and partial characterization of 7,8-dihydro-6-hydroxymethylpterin-pyrophosphokinase and ...
|
|
37540
|
Purification and partial characterization of 7,8-dihydro-6-hydroxymethylpterin-pyrophosphokinase and ...
|
|
37541
|
The hydroxymethyldihydropterin pyrophosphokinase domain of the multifunctional folic acid synthesis Fas ...
|
|
37542
|
Purification and properties of methylamine dehydrogenase from Paracoccus denitrificans
|
|
37543
|
Purification and properties of methylamine dehydrogenase from Paracoccus denitrificans
|
|
37544
|
Biosynthesis of N-glycolylneuraminic acid-containing glycoconjugates. Purification and characterization of the ...
|
|
37545
|
Characterization of undecaprenol kinase from Lactobacillus plantarum
|
|
37546
|
Characterization of undecaprenol kinase from Lactobacillus plantarum
|
|
37547
|
Characterization of undecaprenol kinase from Lactobacillus plantarum
|
|
37548
|
Glycoprotein enzymes secreted by Aspergillus niger: purification and properties of alpha-glaactosidase
|
|
37549
|
Glycoprotein enzymes secreted by Aspergillus niger: purification and properties of alpha-glaactosidase
|
|
37550
|
Purification and properties of the glutathione reductase of Chromatium vinosum
|
|
37551
|
Purification and properties of the glutathione reductase of Chromatium vinosum
|
|
37552
|
Purification and properties of the glutathione reductase of Chromatium vinosum
|
|
37553
|
Purification and properties of the glutathione reductase of Chromatium vinosum
|
|
37554
|
Purification and properties of the glutathione reductase of Chromatium vinosum
|
|
37555
|
Purification and properties of the glutathione reductase of Chromatium vinosum
|
|
37556
|
Purification and properties of the glutathione reductase of Chromatium vinosum
|
|
37557
|
Purification and properties of the glutathione reductase of Chromatium vinosum
|
|
37558
|
Purification and properties of the glutathione reductase of Chromatium vinosum
|
|
37559
|
Purification and characterization of 7 alpha-hydroxy-4-cholesten-3-one 12 alpha-monooxygenase
|
|
37560
|
A monofunctional and thermostable prephenate dehydratase from the archaeon Methanocaldococcus jannaschii
|
|
37561
|
A monofunctional and thermostable prephenate dehydratase from the archaeon Methanocaldococcus jannaschii
|
|
37562
|
Expression and characterization of Lactobacillus 30a histidine decarboxylase in Escherichia coli
|
|
37563
|
Expression and characterization of Lactobacillus 30a histidine decarboxylase in Escherichia coli
|
|
37564
|
Methanococcus jannaschii uses a pyruvoyl-dependent arginine decarboxylase in polyamine biosynthesis
|
|
37565
|
Methanococcus jannaschii uses a pyruvoyl-dependent arginine decarboxylase in polyamine biosynthesis
|
|
37566
|
Methanococcus jannaschii uses a pyruvoyl-dependent arginine decarboxylase in polyamine biosynthesis
|
|
37567
|
Methanococcus jannaschii uses a pyruvoyl-dependent arginine decarboxylase in polyamine biosynthesis
|
|
37568
|
Methanococcus jannaschii uses a pyruvoyl-dependent arginine decarboxylase in polyamine biosynthesis
|
|
37569
|
Detection of CMP-N-acetylneuraminic acid hydroxylase activity in fractionated mouse liver
|
|
37570
|
Aromatic amine dehydrogenase, a second tryptophan tryptophylquinone enzyme
|
|
37571
|
Aromatic amine dehydrogenase, a second tryptophan tryptophylquinone enzyme
|
|
37572
|
Aromatic amine dehydrogenase, a second tryptophan tryptophylquinone enzyme
|
|
37573
|
Aromatic amine dehydrogenase, a second tryptophan tryptophylquinone enzyme
|
|
37574
|
Aromatic amine dehydrogenase, a second tryptophan tryptophylquinone enzyme
|
|
37575
|
Biosynthetic arginine decarboxylase from Escherichia coli. Purification and properties
|
|
37576
|
Biosynthetic arginine decarboxylase from Escherichia coli. Purification and properties
|
|
37577
|
Biosynthetic arginine decarboxylase from Escherichia coli. Purification and properties
|
|
37578
|
Biosynthetic arginine decarboxylase from Escherichia coli. Purification and properties
|
|
37579
|
Biosynthetic arginine decarboxylase from Escherichia coli. Purification and properties
|
|
37580
|
Biosynthetic arginine decarboxylase from Escherichia coli. Purification and properties
|
|
37581
|
Biosynthetic arginine decarboxylase from Escherichia coli. Purification and properties
|
|
37582
|
Biosynthetic arginine decarboxylase from Escherichia coli. Purification and properties
|
|
37583
|
Cloning, expression and characterization of YSA1H, a human adenosine 5'-diphosphosugar pyrophosphatase ...
|
|
37584
|
Cloning, expression and characterization of YSA1H, a human adenosine 5'-diphosphosugar pyrophosphatase ...
|
|
37585
|
Purification and characterization of acetylcoenzyme A: deacetylvindoline 4-O-acetyltransferase from ...
|
|
37586
|
Purification and characterization of acetylcoenzyme A: deacetylvindoline 4-O-acetyltransferase from ...
|
|
37587
|
Purification and characterization of acetylcoenzyme A: deacetylvindoline 4-O-acetyltransferase from ...
|
|
37588
|
Purification and characterization of acetylcoenzyme A: deacetylvindoline 4-O-acetyltransferase from ...
|
|
37589
|
Characterization of the yeast DGK1-encoded CTP-dependent diacylglycerol kinase
|
|
37590
|
Characterization of the yeast DGK1-encoded CTP-dependent diacylglycerol kinase
|
|
37591
|
Characterization of the yeast DGK1-encoded CTP-dependent diacylglycerol kinase
|
|
37592
|
Characterization of the yeast DGK1-encoded CTP-dependent diacylglycerol kinase
|
|
37593
|
Characterization of the yeast DGK1-encoded CTP-dependent diacylglycerol kinase
|
|
37594
|
Mouse AKR1E1 is an ortholog of pig liver NADPH dependent 1,5-anhydro-D-fructose reductase
|
|
37595
|
Identification of a novel lysophospholipid acyltransferase in Saccharomyces cerevisiae
|
|
37596
|
Identification of a novel lysophospholipid acyltransferase in Saccharomyces cerevisiae
|
|
37597
|
Identification of a novel lysophospholipid acyltransferase in Saccharomyces cerevisiae
|
|
37598
|
Identification of a novel lysophospholipid acyltransferase in Saccharomyces cerevisiae
|
|
37599
|
Identification of a novel lysophospholipid acyltransferase in Saccharomyces cerevisiae
|
|
37600
|
tRNA splicing in yeast and wheat germ. A cyclic phosphodiesterase implicated in the metabolism of ADP-ribose ...
|
|
37601
|
tRNA splicing in yeast and wheat germ. A cyclic phosphodiesterase implicated in the metabolism of ADP-ribose ...
|
|
37602
|
tRNA splicing in yeast and wheat germ. A cyclic phosphodiesterase implicated in the metabolism of ADP-ribose ...
|
|
37603
|
tRNA splicing in yeast and wheat germ. A cyclic phosphodiesterase implicated in the metabolism of ADP-ribose ...
|
|
37604
|
tRNA splicing in yeast and wheat germ. A cyclic phosphodiesterase implicated in the metabolism of ADP-ribose ...
|
|
37605
|
tRNA splicing in yeast and wheat germ. A cyclic phosphodiesterase implicated in the metabolism of ADP-ribose ...
|
|
37606
|
tRNA splicing in yeast and wheat germ. A cyclic phosphodiesterase implicated in the metabolism of ADP-ribose ...
|
|
37607
|
tRNA splicing in yeast and wheat germ. A cyclic phosphodiesterase implicated in the metabolism of ADP-ribose ...
|
|
37608
|
tRNA splicing in yeast and wheat germ. A cyclic phosphodiesterase implicated in the metabolism of ADP-ribose ...
|
|
37609
|
tRNA splicing in yeast and wheat germ. A cyclic phosphodiesterase implicated in the metabolism of ADP-ribose ...
|
|
37610
|
tRNA splicing in yeast and wheat germ. A cyclic phosphodiesterase implicated in the metabolism of ADP-ribose ...
|
|
37611
|
tRNA splicing in yeast and wheat germ. A cyclic phosphodiesterase implicated in the metabolism of ADP-ribose ...
|
|
37612
|
tRNA splicing in yeast and wheat germ. A cyclic phosphodiesterase implicated in the metabolism of ADP-ribose ...
|
|
37613
|
tRNA splicing in yeast and wheat germ. A cyclic phosphodiesterase implicated in the metabolism of ADP-ribose ...
|
|
37614
|
Deoxyribose aldolase from Lactobacillus plantarum
|
|
37615
|
Deoxyribose aldolase from Lactobacillus plantarum
|
|
37616
|
Deoxyribose aldolase from Lactobacillus plantarum
|
|
37617
|
Deoxyribose aldolase from Lactobacillus plantarum
|
|
37618
|
Escherichia coli flavohemoglobin is an efficient alkylhydroperoxide reductase
|
|
37619
|
Escherichia coli flavohemoglobin is an efficient alkylhydroperoxide reductase
|
|
37620
|
Escherichia coli flavohemoglobin is an efficient alkylhydroperoxide reductase
|
|
37621
|
Escherichia coli flavohemoglobin is an efficient alkylhydroperoxide reductase
|
|
37622
|
Escherichia coli flavohemoglobin is an efficient alkylhydroperoxide reductase
|
|
37623
|
Escherichia coli flavohemoglobin is an efficient alkylhydroperoxide reductase
|
|
37624
|
Glycolysis_91d1d92c_43c9_4d55_ba43_d8204b37de8f
|
|
37625
|
Glycolysis_5509f4f9_e245_4726_bb19_c544e165e637
|
|
37626
|
Glycolysis_19894d84_0d53_414d_910d_9c8f4f23f463
|
|
37627
|
Glycolysis_1be26fb5_210b_441b_917a_b67ca805deaa
|
|
37628
|
Glycolysis_752024f1_2c9d_4736_a6b5_f70a7f6dac3c
|
|
37629
|
Glycolysis_b697b335_9a83_4db7_9baf_7fa5ae56468d
|
|
37630
|
The coenzyme a biosynthetic enzyme phosphopantetheine adenylyltransferase plays a crucial role in plant ...
|
|
37631
|
The coenzyme a biosynthetic enzyme phosphopantetheine adenylyltransferase plays a crucial role in plant ...
|
|
37632
|
Glycolysis_d5fb6839_2363_4b6a_ae60_9d2a449438ac
|
|
37633
|
Glycolysis_f5a5019c_e560_4c15_aca8_528cf000bca3
|
|
37634
|
Glycolysis_56cd089b_39f2_4d51_b5c2_e8c054a4d747
|
|
37635
|
Glycolysis_23097585_6289_42e8_803f_89a920fd77f5
|
|
37636
|
Glycolysis_4b1b8e58_cde1_4aaf_884e_b797354f3335
|
|
37637
|
Purine biosynthesis pathway_f8426967_72e0_45e7_9eaa_5a896ef9f6a2
|
|
37638
|
Purine biosynthesis pathway_cd8204c7_f891_46af_91dd_b2d0ab191511
|
|
37639
|
Purine biosynthesis pathway_a9ebddc8_e898_424e_9446_1fff5aa6da6b
|
|
37640
|
Purine biosynthesis pathway_c418d95a_5e4e_469f_bdf3_dc59a7c72ec4
|
|
37642
|
Structural and kinetic differences between human and Aspergillus fumigatus D-glucosamine-6-phosphate ...
|
|
37643
|
Structural and kinetic differences between human and Aspergillus fumigatus D-glucosamine-6-phosphate ...
|
|
37644
|
Structural and kinetic differences between human and Aspergillus fumigatus D-glucosamine-6-phosphate ...
|
|
37645
|
Structural and kinetic differences between human and Aspergillus fumigatus D-glucosamine-6-phosphate ...
|
|
37646
|
Structural and kinetic differences between human and Aspergillus fumigatus D-glucosamine-6-phosphate ...
|
|
37647
|
Structural and kinetic differences between human and Aspergillus fumigatus D-glucosamine-6-phosphate ...
|
|
37648
|
Structural and kinetic differences between human and Aspergillus fumigatus D-glucosamine-6-phosphate ...
|
|
37649
|
Structural and kinetic differences between human and Aspergillus fumigatus D-glucosamine-6-phosphate ...
|
|
37650
|
Structural and kinetic differences between human and Aspergillus fumigatus D-glucosamine-6-phosphate ...
|
|
37651
|
Structural and kinetic differences between human and Aspergillus fumigatus D-glucosamine-6-phosphate ...
|
|
37652
|
Structural and kinetic differences between human and Aspergillus fumigatus D-glucosamine-6-phosphate ...
|
|
37653
|
Structural and kinetic differences between human and Aspergillus fumigatus D-glucosamine-6-phosphate ...
|
|
37654
|
Purification and biochemical characterization of pyruvate oxidase from Lactobacillus plantarum
|
|
37655
|
Purification and biochemical characterization of pyruvate oxidase from Lactobacillus plantarum
|
|
37656
|
Purification and biochemical characterization of pyruvate oxidase from Lactobacillus plantarum
|
|
37657
|
Molecular, Kinetic, and Immunological Properties of the 6-Phosphofructokinase from the Green Alga Selenastrum ...
|
|
37658
|
Molecular, Kinetic, and Immunological Properties of the 6-Phosphofructokinase from the Green Alga Selenastrum ...
|
|
37659
|
Molecular, Kinetic, and Immunological Properties of the 6-Phosphofructokinase from the Green Alga Selenastrum ...
|
|
37660
|
Molecular, Kinetic, and Immunological Properties of the 6-Phosphofructokinase from the Green Alga Selenastrum ...
|
|
37661
|
Molecular, Kinetic, and Immunological Properties of the 6-Phosphofructokinase from the Green Alga Selenastrum ...
|
|
37662
|
Molecular, Kinetic, and Immunological Properties of the 6-Phosphofructokinase from the Green Alga Selenastrum ...
|
|
37663
|
Molecular, Kinetic, and Immunological Properties of the 6-Phosphofructokinase from the Green Alga Selenastrum ...
|
|
37664
|
Molecular, Kinetic, and Immunological Properties of the 6-Phosphofructokinase from the Green Alga Selenastrum ...
|
|
37665
|
Molecular, Kinetic, and Immunological Properties of the 6-Phosphofructokinase from the Green Alga Selenastrum ...
|
|
37666
|
Molecular, Kinetic, and Immunological Properties of the 6-Phosphofructokinase from the Green Alga Selenastrum ...
|
|
37667
|
Molecular, Kinetic, and Immunological Properties of the 6-Phosphofructokinase from the Green Alga Selenastrum ...
|
|
37668
|
Molecular, Kinetic, and Immunological Properties of the 6-Phosphofructokinase from the Green Alga Selenastrum ...
|
|
37669
|
Molecular, Kinetic, and Immunological Properties of the 6-Phosphofructokinase from the Green Alga Selenastrum ...
|
|
37670
|
Molecular, Kinetic, and Immunological Properties of the 6-Phosphofructokinase from the Green Alga Selenastrum ...
|
|
37671
|
Molecular, Kinetic, and Immunological Properties of the 6-Phosphofructokinase from the Green Alga Selenastrum ...
|
|
37672
|
Molecular, Kinetic, and Immunological Properties of the 6-Phosphofructokinase from the Green Alga Selenastrum ...
|
|
37673
|
Reaction path of phosphofructo-1-kinase is altered by mutagenesis and alternative substrates
|
|
37674
|
Reaction path of phosphofructo-1-kinase is altered by mutagenesis and alternative substrates
|
|
37675
|
Reaction path of phosphofructo-1-kinase is altered by mutagenesis and alternative substrates
|
|
37676
|
Reaction path of phosphofructo-1-kinase is altered by mutagenesis and alternative substrates
|
|
37677
|
Reaction path of phosphofructo-1-kinase is altered by mutagenesis and alternative substrates
|
|
37678
|
Reaction path of phosphofructo-1-kinase is altered by mutagenesis and alternative substrates
|
|
37679
|
Reaction path of phosphofructo-1-kinase is altered by mutagenesis and alternative substrates
|
|
37680
|
Reaction path of phosphofructo-1-kinase is altered by mutagenesis and alternative substrates
|
|
37681
|
Reaction path of phosphofructo-1-kinase is altered by mutagenesis and alternative substrates
|
|
37682
|
Reaction path of phosphofructo-1-kinase is altered by mutagenesis and alternative substrates
|
|
37683
|
Reaction path of phosphofructo-1-kinase is altered by mutagenesis and alternative substrates
|
|
37684
|
Purification and comparison of sex-dependent and sex-independent gulonolactonase of rats
|
|
37685
|
Purification and comparison of sex-dependent and sex-independent gulonolactonase of rats
|
|
37686
|
Purification and comparison of sex-dependent and sex-independent gulonolactonase of rats
|
|
37687
|
Purification and comparison of sex-dependent and sex-independent gulonolactonase of rats
|
|
37688
|
Purification and comparison of sex-dependent and sex-independent gulonolactonase of rats
|
|
37689
|
Purification and comparison of sex-dependent and sex-independent gulonolactonase of rats
|
|
37690
|
Purification and comparison of sex-dependent and sex-independent gulonolactonase of rats
|
|
37691
|
Purification and comparison of sex-dependent and sex-independent gulonolactonase of rats
|
|
37692
|
Paramecium bursaria chlorella virus-1 encodes an unusual arginine decarboxylase that is a close homolog of ...
|
|
37693
|
Paramecium bursaria chlorella virus-1 encodes an unusual arginine decarboxylase that is a close homolog of ...
|
|
37694
|
Paramecium bursaria chlorella virus-1 encodes an unusual arginine decarboxylase that is a close homolog of ...
|
|
37695
|
Paramecium bursaria chlorella virus-1 encodes an unusual arginine decarboxylase that is a close homolog of ...
|
|
37696
|
Paramecium bursaria chlorella virus-1 encodes an unusual arginine decarboxylase that is a close homolog of ...
|
|
37697
|
Paramecium bursaria chlorella virus-1 encodes an unusual arginine decarboxylase that is a close homolog of ...
|
|
37698
|
Paramecium bursaria chlorella virus-1 encodes an unusual arginine decarboxylase that is a close homolog of ...
|
|
37699
|
Paramecium bursaria chlorella virus-1 encodes an unusual arginine decarboxylase that is a close homolog of ...
|
|
37700
|
Paramecium bursaria chlorella virus-1 encodes an unusual arginine decarboxylase that is a close homolog of ...
|
|
37701
|
Paramecium bursaria chlorella virus-1 encodes an unusual arginine decarboxylase that is a close homolog of ...
|
|
37702
|
Paramecium bursaria chlorella virus-1 encodes an unusual arginine decarboxylase that is a close homolog of ...
|
|
37703
|
Kinetic characteristics of phosphofructokinase from Bacillus stearothermophilus: MgATP nonallosterically ...
|
|
37704
|
Kinetic characteristics of phosphofructokinase from Bacillus stearothermophilus: MgATP nonallosterically ...
|
|
37705
|
Kinetic characteristics of phosphofructokinase from Bacillus stearothermophilus: MgATP nonallosterically ...
|
|
37706
|
Kinetic characteristics of phosphofructokinase from Bacillus stearothermophilus: MgATP nonallosterically ...
|
|
37707
|
Kinetic characteristics of phosphofructokinase from Bacillus stearothermophilus: MgATP nonallosterically ...
|
|
37708
|
Kinetic characteristics of phosphofructokinase from Bacillus stearothermophilus: MgATP nonallosterically ...
|
|
37709
|
Kinetic characteristics of phosphofructokinase from Bacillus stearothermophilus: MgATP nonallosterically ...
|
|
37710
|
Kinetic characteristics of phosphofructokinase from Bacillus stearothermophilus: MgATP nonallosterically ...
|
|
37711
|
Kinetic characteristics of phosphofructokinase from Bacillus stearothermophilus: MgATP nonallosterically ...
|
|
37712
|
Kinetic characteristics of phosphofructokinase from Bacillus stearothermophilus: MgATP nonallosterically ...
|
|
37713
|
Kinetic characteristics of phosphofructokinase from Bacillus stearothermophilus: MgATP nonallosterically ...
|
|
37714
|
Kinetic characteristics of phosphofructokinase from Bacillus stearothermophilus: MgATP nonallosterically ...
|
|
37715
|
Kinetic characteristics of phosphofructokinase from Bacillus stearothermophilus: MgATP nonallosterically ...
|
|
37716
|
Kinetic characteristics of phosphofructokinase from Bacillus stearothermophilus: MgATP nonallosterically ...
|
|
37717
|
Kinetic characteristics of phosphofructokinase from Bacillus stearothermophilus: MgATP nonallosterically ...
|
|
37718
|
Kinetic characteristics of phosphofructokinase from Bacillus stearothermophilus: MgATP nonallosterically ...
|
|
37719
|
Kinetic characteristics of phosphofructokinase from Bacillus stearothermophilus: MgATP nonallosterically ...
|
|
37720
|
Kinetic characteristics of phosphofructokinase from Bacillus stearothermophilus: MgATP nonallosterically ...
|
|
37721
|
Kinetic characteristics of phosphofructokinase from Bacillus stearothermophilus: MgATP nonallosterically ...
|
|
37722
|
Kinetic characteristics of phosphofructokinase from Bacillus stearothermophilus: MgATP nonallosterically ...
|
|
37723
|
Kinetic characteristics of phosphofructokinase from Bacillus stearothermophilus: MgATP nonallosterically ...
|
|
37724
|
Kinetic characteristics of phosphofructokinase from Bacillus stearothermophilus: MgATP nonallosterically ...
|
|
37725
|
Kinetic characteristics of phosphofructokinase from Bacillus stearothermophilus: MgATP nonallosterically ...
|
|
37726
|
Kinetic characteristics of phosphofructokinase from Bacillus stearothermophilus: MgATP nonallosterically ...
|
|
37727
|
Identifying latent enzyme activities: substrate ambiguity within modern bacterial sugar kinases
|
|
37728
|
Identifying latent enzyme activities: substrate ambiguity within modern bacterial sugar kinases
|
|
37729
|
Identifying latent enzyme activities: substrate ambiguity within modern bacterial sugar kinases
|
|
37730
|
Identifying latent enzyme activities: substrate ambiguity within modern bacterial sugar kinases
|
|
37731
|
Identifying latent enzyme activities: substrate ambiguity within modern bacterial sugar kinases
|
|
37732
|
Identifying latent enzyme activities: substrate ambiguity within modern bacterial sugar kinases
|
|
37733
|
Identifying latent enzyme activities: substrate ambiguity within modern bacterial sugar kinases
|
|
37734
|
Identifying latent enzyme activities: substrate ambiguity within modern bacterial sugar kinases
|
|
37735
|
Identifying latent enzyme activities: substrate ambiguity within modern bacterial sugar kinases
|
|
37736
|
Identifying latent enzyme activities: substrate ambiguity within modern bacterial sugar kinases
|
|
37737
|
Identifying latent enzyme activities: substrate ambiguity within modern bacterial sugar kinases
|
|
37738
|
Identifying latent enzyme activities: substrate ambiguity within modern bacterial sugar kinases
|
|
37739
|
NADPH-cytochrome c reductase in outer membrane of kidney mitochondria. Purification and properties
|
|
37740
|
NADPH-cytochrome c reductase in outer membrane of kidney mitochondria. Purification and properties
|
|
37741
|
NADPH-cytochrome c reductase in outer membrane of kidney mitochondria. Purification and properties
|
|
37742
|
NADPH-cytochrome c reductase in outer membrane of kidney mitochondria. Purification and properties
|
|
37743
|
NADPH-cytochrome c reductase in outer membrane of kidney mitochondria. Purification and properties
|
|
37744
|
NADPH-cytochrome c reductase in outer membrane of kidney mitochondria. Purification and properties
|
|
37745
|
Glucose-induced conformational changes in glucokinase mediate allosteric regulation: transient kinetic ...
|
|
37746
|
Glucose-induced conformational changes in glucokinase mediate allosteric regulation: transient kinetic ...
|
|
37747
|
Glucose-induced conformational changes in glucokinase mediate allosteric regulation: transient kinetic ...
|
|
37748
|
Glucose-induced conformational changes in glucokinase mediate allosteric regulation: transient kinetic ...
|
|
37749
|
Glucose-induced conformational changes in glucokinase mediate allosteric regulation: transient kinetic ...
|
|
37750
|
Glucose-induced conformational changes in glucokinase mediate allosteric regulation: transient kinetic ...
|
|
37751
|
Glucose-induced conformational changes in glucokinase mediate allosteric regulation: transient kinetic ...
|
|
37752
|
Glucose-induced conformational changes in glucokinase mediate allosteric regulation: transient kinetic ...
|
|
37753
|
Glucose-induced conformational changes in glucokinase mediate allosteric regulation: transient kinetic ...
|
|
37754
|
Glucose-induced conformational changes in glucokinase mediate allosteric regulation: transient kinetic ...
|
|
37755
|
Glucose-induced conformational changes in glucokinase mediate allosteric regulation: transient kinetic ...
|
|
37756
|
Glucose-induced conformational changes in glucokinase mediate allosteric regulation: transient kinetic ...
|
|
37757
|
Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening
|
|
37758
|
Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening
|
|
37759
|
Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening
|
|
37760
|
Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening
|
|
37761
|
Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening
|
|
37762
|
Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening
|
|
37763
|
Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening
|
|
37764
|
Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening
|
|
37765
|
Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening
|
|
37766
|
Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening
|
|
37767
|
Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening
|
|
37768
|
Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening
|
|
37769
|
The novel tungsten-iron-sulfur protein of the hyperthermophilic archaebacterium, Pyrococcus furiosus, is an ...
|
|
37770
|
The novel tungsten-iron-sulfur protein of the hyperthermophilic archaebacterium, Pyrococcus furiosus, is an ...
|
|
37771
|
The novel tungsten-iron-sulfur protein of the hyperthermophilic archaebacterium, Pyrococcus furiosus, is an ...
|
|
37772
|
The novel tungsten-iron-sulfur protein of the hyperthermophilic archaebacterium, Pyrococcus furiosus, is an ...
|
|
37773
|
The novel tungsten-iron-sulfur protein of the hyperthermophilic archaebacterium, Pyrococcus furiosus, is an ...
|
|
37774
|
The novel tungsten-iron-sulfur protein of the hyperthermophilic archaebacterium, Pyrococcus furiosus, is an ...
|
|
37775
|
The human homolog of the rat inositol phosphate multikinase is an inositol 1,3,4,6-tetrakisphosphate 5-kinase
|
|
37776
|
The human homolog of the rat inositol phosphate multikinase is an inositol 1,3,4,6-tetrakisphosphate 5-kinase
|
|
37777
|
The human homolog of the rat inositol phosphate multikinase is an inositol 1,3,4,6-tetrakisphosphate 5-kinase
|
|
37787
|
Characterization of a benzyl alcohol dehydrogenase from Lactobacillus plantarum WCFS1
|
|
37788
|
The metabolism of L-fucose. IV. The biosynthesis of guanosine diphosphate L-fucose in porcine liver
|
|
37789
|
Phosphomevalonate Kinase: Functional Investigation of the Recombinant Human Enzyme
|
|
37790
|
Phosphomevalonate Kinase: Functional Investigation of the Recombinant Human Enzyme
|
|
37791
|
Phosphomevalonate Kinase: Functional Investigation of the Recombinant Human Enzyme
|
|
37792
|
Phosphomevalonate Kinase: Functional Investigation of the Recombinant Human Enzyme
|
|
37793
|
Phosphomevalonate Kinase: Functional Investigation of the Recombinant Human Enzyme
|
|
37794
|
Phosphomevalonate Kinase: Functional Investigation of the Recombinant Human Enzyme
|
|
37795
|
Phosphomevalonate Kinase: Functional Investigation of the Recombinant Human Enzyme
|
|
37796
|
Phosphomevalonate Kinase: Functional Investigation of the Recombinant Human Enzyme
|
|
37797
|
Phosphomevalonate Kinase: Functional Investigation of the Recombinant Human Enzyme
|
|
37798
|
Phosphomevalonate Kinase: Functional Investigation of the Recombinant Human Enzyme
|
|
37799
|
Phosphomevalonate Kinase: Functional Investigation of the Recombinant Human Enzyme
|
|
37800
|
Phosphomevalonate Kinase: Functional Investigation of the Recombinant Human Enzyme
|
|
37801
|
Phosphomevalonate Kinase: Functional Investigation of the Recombinant Human Enzyme
|
|
37802
|
Phosphomevalonate Kinase: Functional Investigation of the Recombinant Human Enzyme
|
|
37803
|
Phosphomevalonate Kinase: Functional Investigation of the Recombinant Human Enzyme
|
|
37804
|
Phosphomevalonate Kinase: Functional Investigation of the Recombinant Human Enzyme
|
|
37805
|
Phosphomevalonate Kinase: Functional Investigation of the Recombinant Human Enzyme
|
|
37806
|
Phosphomevalonate Kinase: Functional Investigation of the Recombinant Human Enzyme
|
|
37807
|
Phosphomevalonate Kinase: Functional Investigation of the Recombinant Human Enzyme
|
|
37808
|
Phosphomevalonate Kinase: Functional Investigation of the Recombinant Human Enzyme
|
|
37809
|
Phosphomevalonate Kinase: Functional Investigation of the Recombinant Human Enzyme
|
|
37810
|
Phosphomevalonate Kinase: Functional Investigation of the Recombinant Human Enzyme
|
|
37811
|
Identification and cloning of a gene encoding tannase (tannin acylhydrolase) from Lactobacillus plantarum ATCC ...
|
|
37812
|
Identification and cloning of a gene encoding tannase (tannin acylhydrolase) from Lactobacillus plantarum ATCC ...
|
|
37813
|
Identification and cloning of a gene encoding tannase (tannin acylhydrolase) from Lactobacillus plantarum ATCC ...
|
|
37814
|
Anti-influenza prodrug oseltamivir is activated by carboxylesterase human carboxylesterase 1, and the ...
|
|
37815
|
Anti-influenza prodrug oseltamivir is activated by carboxylesterase human carboxylesterase 1, and the ...
|
|
37816
|
Characterization of an l-arabinose isomerase from the Lactobacillus plantarum NC8 strain showing pronounced ...
|
|
37817
|
Characterization of an l-arabinose isomerase from the Lactobacillus plantarum NC8 strain showing pronounced ...
|
|
37818
|
Identification of two active site residues in human angiotensin I-converting enzyme.
|
|
37819
|
Identification of two active site residues in human angiotensin I-converting enzyme.
|
|
37820
|
Identification of two active site residues in human angiotensin I-converting enzyme.
|
|
37821
|
Identification of two active site residues in human angiotensin I-converting enzyme.
|
|
37822
|
Identification of two active site residues in human angiotensin I-converting enzyme.
|
|
37823
|
Identification of two active site residues in human angiotensin I-converting enzyme.
|
|
37824
|
Identification of two active site residues in human angiotensin I-converting enzyme.
|
|
37825
|
Identification of two active site residues in human angiotensin I-converting enzyme.
|
|
37826
|
Identification of two active site residues in human angiotensin I-converting enzyme.
|
|
37827
|
A new nicotinamide-adenine dinucleotide-dependent hydroaromatic dehydrogenase of Lactobacillus plantarum and ...
|
|
37828
|
A new nicotinamide-adenine dinucleotide-dependent hydroaromatic dehydrogenase of Lactobacillus plantarum and ...
|
|
37829
|
A new nicotinamide-adenine dinucleotide-dependent hydroaromatic dehydrogenase of Lactobacillus plantarum and ...
|
|
37830
|
A new nicotinamide-adenine dinucleotide-dependent hydroaromatic dehydrogenase of Lactobacillus plantarum and ...
|
|
37831
|
A new nicotinamide-adenine dinucleotide-dependent hydroaromatic dehydrogenase of Lactobacillus plantarum and ...
|
|
37832
|
Undecaprenyl pyrophosphate synthetase from Lactobacillus plantarum: a dimeric protein
|
|
37833
|
Undecaprenyl pyrophosphate synthetase from Lactobacillus plantarum: a dimeric protein
|
|
37834
|
Undecaprenyl pyrophosphate synthetase from Lactobacillus plantarum: a dimeric protein
|
|
37835
|
Undecaprenyl pyrophosphate synthetase from Lactobacillus plantarum: a dimeric protein
|
|
37836
|
Undecaprenyl pyrophosphate synthetase from Lactobacillus plantarum: a dimeric protein
|
|
37837
|
Undecaprenyl pyrophosphate synthetase from Lactobacillus plantarum: a dimeric protein
|
|
37838
|
Undecaprenyl pyrophosphate synthetase from Lactobacillus plantarum: a dimeric protein
|
|
37839
|
Undecaprenyl pyrophosphate synthetase from Lactobacillus plantarum: a dimeric protein
|
|
37840
|
Undecaprenyl pyrophosphate synthetase from Lactobacillus plantarum: a dimeric protein
|
|
37841
|
Undecaprenyl pyrophosphate synthetase from Lactobacillus plantarum: a dimeric protein
|
|
37842
|
Undecaprenyl pyrophosphate synthetase from Lactobacillus plantarum: a dimeric protein
|
|
37843
|
Undecaprenyl pyrophosphate synthetase from Lactobacillus plantarum: a dimeric protein
|
|
37844
|
Undecaprenyl pyrophosphate synthetase from Lactobacillus plantarum: a dimeric protein
|
|
37845
|
Functional Expression of Two Arabidopsis Aldehyde Oxidases in the Yeast Pichia pastoris
|
|
37846
|
Functional Expression of Two Arabidopsis Aldehyde Oxidases in the Yeast Pichia pastoris
|
|
37847
|
Functional Expression of Two Arabidopsis Aldehyde Oxidases in the Yeast Pichia pastoris
|
|
37848
|
Functional Expression of Two Arabidopsis Aldehyde Oxidases in the Yeast Pichia pastoris
|
|
37849
|
Functional Expression of Two Arabidopsis Aldehyde Oxidases in the Yeast Pichia pastoris
|
|
37850
|
Functional Expression of Two Arabidopsis Aldehyde Oxidases in the Yeast Pichia pastoris
|
|
37851
|
Functional Expression of Two Arabidopsis Aldehyde Oxidases in the Yeast Pichia pastoris
|
|
37852
|
Functional Expression of Two Arabidopsis Aldehyde Oxidases in the Yeast Pichia pastoris
|
|
37853
|
Functional Expression of Two Arabidopsis Aldehyde Oxidases in the Yeast Pichia pastoris
|
|
37854
|
Functional Expression of Two Arabidopsis Aldehyde Oxidases in the Yeast Pichia pastoris
|
|
37855
|
Functional Expression of Two Arabidopsis Aldehyde Oxidases in the Yeast Pichia pastoris
|
|
37856
|
Inhibition of polymorphic human carbonyl reductase 1 (CBR1) by the cardioprotectant flavonoid ...
|
|
37857
|
Inhibition of polymorphic human carbonyl reductase 1 (CBR1) by the cardioprotectant flavonoid ...
|
|
37858
|
Inhibition of polymorphic human carbonyl reductase 1 (CBR1) by the cardioprotectant flavonoid ...
|
|
37859
|
Inhibition of polymorphic human carbonyl reductase 1 (CBR1) by the cardioprotectant flavonoid ...
|
|
37860
|
Inhibition of polymorphic human carbonyl reductase 1 (CBR1) by the cardioprotectant flavonoid ...
|
|
37861
|
Inhibition of polymorphic human carbonyl reductase 1 (CBR1) by the cardioprotectant flavonoid ...
|
|
37862
|
Inhibition of polymorphic human carbonyl reductase 1 (CBR1) by the cardioprotectant flavonoid ...
|
|
37863
|
Inhibition of polymorphic human carbonyl reductase 1 (CBR1) by the cardioprotectant flavonoid ...
|
|
37864
|
Inhibition of polymorphic human carbonyl reductase 1 (CBR1) by the cardioprotectant flavonoid ...
|
|
37865
|
Inhibition of polymorphic human carbonyl reductase 1 (CBR1) by the cardioprotectant flavonoid ...
|
|
37866
|
Inhibition of polymorphic human carbonyl reductase 1 (CBR1) by the cardioprotectant flavonoid ...
|
|
37867
|
Inhibition of polymorphic human carbonyl reductase 1 (CBR1) by the cardioprotectant flavonoid ...
|
|
37868
|
Inhibition of polymorphic human carbonyl reductase 1 (CBR1) by the cardioprotectant flavonoid ...
|
|
37869
|
Inhibition of polymorphic human carbonyl reductase 1 (CBR1) by the cardioprotectant flavonoid ...
|
|
37870
|
Inhibition of polymorphic human carbonyl reductase 1 (CBR1) by the cardioprotectant flavonoid ...
|
|
37871
|
Inhibition of polymorphic human carbonyl reductase 1 (CBR1) by the cardioprotectant flavonoid ...
|
|
37872
|
Inhibition of polymorphic human carbonyl reductase 1 (CBR1) by the cardioprotectant flavonoid ...
|
|
37873
|
Inhibition of polymorphic human carbonyl reductase 1 (CBR1) by the cardioprotectant flavonoid ...
|
|
37874
|
Dephosphorylation of 1D-myo-inositol 1,4-bisphosphate in rat liver
|
|
37875
|
Dephosphorylation of 1D-myo-inositol 1,4-bisphosphate in rat liver
|
|
37876
|
Dephosphorylation of 1D-myo-inositol 1,4-bisphosphate in rat liver
|
|
37877
|
Dephosphorylation of 1D-myo-inositol 1,4-bisphosphate in rat liver
|
|
37878
|
Dephosphorylation of 1D-myo-inositol 1,4-bisphosphate in rat liver
|
|
37879
|
Dephosphorylation of 1D-myo-inositol 1,4-bisphosphate in rat liver
|
|
37880
|
Dephosphorylation of 1D-myo-inositol 1,4-bisphosphate in rat liver
|
|
37881
|
Dephosphorylation of 1D-myo-inositol 1,4-bisphosphate in rat liver
|
|
37882
|
Dephosphorylation of 1D-myo-inositol 1,4-bisphosphate in rat liver
|
|
37883
|
Dephosphorylation of 1D-myo-inositol 1,4-bisphosphate in rat liver
|
|
37884
|
Dephosphorylation of 1D-myo-inositol 1,4-bisphosphate in rat liver
|
|
37885
|
D-myo-inositol (1,4)-bisphosphate 1-phosphate. Partial purification from rat liver and characterization
|
|
37886
|
Purification and Properties of Kynureninase from Rat Liver
|
|
37887
|
Purification and Properties of Kynureninase from Rat Liver
|
|
37888
|
Purification and Properties of Kynureninase from Rat Liver
|
|
37889
|
Purification and Properties of Kynureninase from Rat Liver
|
|
37890
|
Purification and Properties of Kynureninase from Rat Liver
|
|
37891
|
Purification and Properties of Kynureninase from Rat Liver
|
|
37892
|
Purification and Properties of Kynureninase from Rat Liver
|
|
37893
|
Purification and Properties of Kynureninase from Rat Liver
|
|
37894
|
Methylphenidate is stereoselectively hydrolyzed by human carboxylesterase CES1A1
|
|
37895
|
Methylphenidate is stereoselectively hydrolyzed by human carboxylesterase CES1A1
|
|
37896
|
Methylphenidate is stereoselectively hydrolyzed by human carboxylesterase CES1A1
|
|
37897
|
Methylphenidate is stereoselectively hydrolyzed by human carboxylesterase CES1A1
|
|
37898
|
Methylphenidate is stereoselectively hydrolyzed by human carboxylesterase CES1A1
|
|
37899
|
Methylphenidate is stereoselectively hydrolyzed by human carboxylesterase CES1A1
|
|
37900
|
Methylphenidate is stereoselectively hydrolyzed by human carboxylesterase CES1A1
|
|
37901
|
Methylphenidate is stereoselectively hydrolyzed by human carboxylesterase CES1A1
|
|
37902
|
Characterization of the p-coumaric acid decarboxylase from Lactobacillus plantarum CECT 748(T)
|
|
37903
|
Characterization of the p-coumaric acid decarboxylase from Lactobacillus plantarum CECT 748(T)
|
|
37904
|
Characterization of the p-coumaric acid decarboxylase from Lactobacillus plantarum CECT 748(T)
|
|
37905
|
Identification of sn-glycerol-1-phosphate dehydrogenase activity from genomic information on a ...
|
|
37906
|
Glucose-6-phosphate dehydrogenase. Purification and partial characterization
|
|
37907
|
Purification, characterization and identification of rat liver mitochondrial kynurenine aminotransferase with ...
|
|
37908
|
Purification, characterization and identification of rat liver mitochondrial kynurenine aminotransferase with ...
|
|
37909
|
Purification, characterization and identification of rat liver mitochondrial kynurenine aminotransferase with ...
|
|
37910
|
Purification, characterization and identification of rat liver mitochondrial kynurenine aminotransferase with ...
|
|
37911
|
Purification, characterization and identification of rat liver mitochondrial kynurenine aminotransferase with ...
|
|
37912
|
Purification, characterization and identification of rat liver mitochondrial kynurenine aminotransferase with ...
|
|
37913
|
Purification, characterization and identification of rat liver mitochondrial kynurenine aminotransferase with ...
|
|
37914
|
Purification, characterization and identification of rat liver mitochondrial kynurenine aminotransferase with ...
|
|
37915
|
Purification, characterization and identification of rat liver mitochondrial kynurenine aminotransferase with ...
|
|
37916
|
Purification, characterization and identification of rat liver mitochondrial kynurenine aminotransferase with ...
|
|
37917
|
Purification, characterization and identification of rat liver mitochondrial kynurenine aminotransferase with ...
|
|
37918
|
Purification, characterization and identification of rat liver mitochondrial kynurenine aminotransferase with ...
|
|
37919
|
Purification, characterization and identification of rat liver mitochondrial kynurenine aminotransferase with ...
|
|
37920
|
Purification, characterization and identification of rat liver mitochondrial kynurenine aminotransferase with ...
|
|
37921
|
Purification, characterization and identification of rat liver mitochondrial kynurenine aminotransferase with ...
|
|
37922
|
Purification, characterization and identification of rat liver mitochondrial kynurenine aminotransferase with ...
|
|
37923
|
Purification, characterization and identification of rat liver mitochondrial kynurenine aminotransferase with ...
|
|
37924
|
Purification, characterization and identification of rat liver mitochondrial kynurenine aminotransferase with ...
|
|
37925
|
Purification, characterization and identification of rat liver mitochondrial kynurenine aminotransferase with ...
|
|
37926
|
Purification, characterization and identification of rat liver mitochondrial kynurenine aminotransferase with ...
|
|
37927
|
Crystal structure of human liver Delta4-3-ketosteroid 5beta-reductase (AKR1D1) and implications for substrate ...
|
|
37928
|
A functional genetic polymorphism on human carbonyl reductase 1 (CBR1 V88I) impacts on catalytic activity and ...
|
|
37929
|
A functional genetic polymorphism on human carbonyl reductase 1 (CBR1 V88I) impacts on catalytic activity and ...
|
|
37930
|
A functional genetic polymorphism on human carbonyl reductase 1 (CBR1 V88I) impacts on catalytic activity and ...
|
|
37931
|
A functional genetic polymorphism on human carbonyl reductase 1 (CBR1 V88I) impacts on catalytic activity and ...
|
|
37932
|
A functional genetic polymorphism on human carbonyl reductase 1 (CBR1 V88I) impacts on catalytic activity and ...
|
|
37933
|
A functional genetic polymorphism on human carbonyl reductase 1 (CBR1 V88I) impacts on catalytic activity and ...
|
|
37934
|
A functional genetic polymorphism on human carbonyl reductase 1 (CBR1 V88I) impacts on catalytic activity and ...
|
|
37935
|
A functional genetic polymorphism on human carbonyl reductase 1 (CBR1 V88I) impacts on catalytic activity and ...
|
|
37936
|
A functional genetic polymorphism on human carbonyl reductase 1 (CBR1 V88I) impacts on catalytic activity and ...
|
|
37937
|
A functional genetic polymorphism on human carbonyl reductase 1 (CBR1 V88I) impacts on catalytic activity and ...
|
|
37938
|
A functional genetic polymorphism on human carbonyl reductase 1 (CBR1 V88I) impacts on catalytic activity and ...
|
|
37939
|
A functional genetic polymorphism on human carbonyl reductase 1 (CBR1 V88I) impacts on catalytic activity and ...
|
|
37940
|
The fructose 6-phosphate site of phosphofructokinase. Epimeric specificity
|
|
37941
|
The fructose 6-phosphate site of phosphofructokinase. Epimeric specificity
|
|
37942
|
The fructose 6-phosphate site of phosphofructokinase. Epimeric specificity
|
|
37943
|
The fructose 6-phosphate site of phosphofructokinase. Epimeric specificity
|
|
37944
|
Studies on phosphate-activated 5'-methylthioadenosine nucleosidase from human placenta
|
|
37945
|
Active site of Zn(2+)-dependent sn-glycerol-1-phosphate dehydrogenase from Aeropyrum pernix K1
|
|
37946
|
Active site of Zn(2+)-dependent sn-glycerol-1-phosphate dehydrogenase from Aeropyrum pernix K1
|
|
37947
|
Active site of Zn(2+)-dependent sn-glycerol-1-phosphate dehydrogenase from Aeropyrum pernix K1
|
|
37948
|
Active site of Zn(2+)-dependent sn-glycerol-1-phosphate dehydrogenase from Aeropyrum pernix K1
|
|
37949
|
Active site of Zn(2+)-dependent sn-glycerol-1-phosphate dehydrogenase from Aeropyrum pernix K1
|
|
37950
|
Active site of Zn(2+)-dependent sn-glycerol-1-phosphate dehydrogenase from Aeropyrum pernix K1
|
|
37951
|
Active site of Zn(2+)-dependent sn-glycerol-1-phosphate dehydrogenase from Aeropyrum pernix K1
|
|
37952
|
Active site of Zn(2+)-dependent sn-glycerol-1-phosphate dehydrogenase from Aeropyrum pernix K1
|
|
37953
|
Active site of Zn(2+)-dependent sn-glycerol-1-phosphate dehydrogenase from Aeropyrum pernix K1
|
|
37954
|
Characterization studies of glucose dehydrogenase
|
|
37955
|
Characterization studies of glucose dehydrogenase
|
|
37956
|
Characterization studies of glucose dehydrogenase
|
|
37957
|
Characterization studies of glucose dehydrogenase
|
|
37958
|
Characterization studies of glucose dehydrogenase
|
|
37959
|
Characterization studies of glucose dehydrogenase
|
|
37960
|
Inositol polyphosphate 1-phosphatase from calf brain. Purification and inhibition by Li+, Ca2+, and Mn2+
|
|
37961
|
Inositol polyphosphate 1-phosphatase from calf brain. Purification and inhibition by Li+, Ca2+, and Mn2+
|
|
37962
|
Inositol polyphosphate 1-phosphatase from calf brain. Purification and inhibition by Li+, Ca2+, and Mn2+
|
|
37963
|
Inositol polyphosphate 1-phosphatase from calf brain. Purification and inhibition by Li+, Ca2+, and Mn2+
|
|
37964
|
Inositol polyphosphate 1-phosphatase from calf brain. Purification and inhibition by Li+, Ca2+, and Mn2+
|
|
37965
|
Inositol polyphosphate 1-phosphatase from calf brain. Purification and inhibition by Li+, Ca2+, and Mn2+
|
|
37966
|
Inositol polyphosphate 1-phosphatase from calf brain. Purification and inhibition by Li+, Ca2+, and Mn2+
|
|
37967
|
Inositol polyphosphate 1-phosphatase from calf brain. Purification and inhibition by Li+, Ca2+, and Mn2+
|
|
37968
|
Inositol polyphosphate 1-phosphatase from calf brain. Purification and inhibition by Li+, Ca2+, and Mn2+
|
|
37969
|
Inositol polyphosphate 1-phosphatase from calf brain. Purification and inhibition by Li+, Ca2+, and Mn2+
|
|
37970
|
Mechanism of deoxyadenosine-induced catabolism of adenine ribonucleotides in adenosine deaminase-inhibited ...
|
|
37971
|
Mechanism of deoxyadenosine-induced catabolism of adenine ribonucleotides in adenosine deaminase-inhibited ...
|
|
37972
|
Mechanism of deoxyadenosine-induced catabolism of adenine ribonucleotides in adenosine deaminase-inhibited ...
|
|
37973
|
Mechanism of deoxyadenosine-induced catabolism of adenine ribonucleotides in adenosine deaminase-inhibited ...
|
|
37974
|
Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to ...
|
|
37975
|
Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to ...
|
|
37976
|
Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to ...
|
|
37977
|
Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to ...
|
|
37978
|
Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to ...
|
|
37979
|
Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to ...
|
|
37980
|
Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to ...
|
|
37981
|
Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to ...
|
|
37982
|
Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to ...
|
|
37983
|
Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to ...
|
|
37984
|
Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to ...
|
|
37985
|
Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to ...
|
|
37986
|
Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to ...
|
|
37987
|
Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to ...
|
|
37988
|
Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to ...
|
|
37989
|
Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to ...
|
|
37990
|
Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to ...
|
|
37991
|
Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to ...
|
|
37992
|
Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to ...
|
|
37993
|
Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to ...
|
|
37994
|
Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to ...
|
|
37995
|
Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to ...
|
|
37996
|
Glucose dehydrogenase from pig liver. I. Isolation and purification
|
|
37997
|
Glucose dehydrogenase from pig liver. I. Isolation and purification
|
|
37998
|
Lipid activation of undecaprenol kinase from Lactobacillus plantarum
|
|
37999
|
Lipid activation of undecaprenol kinase from Lactobacillus plantarum
|
|
38000
|
Purification and properties of rat liver fructokinase
|





 If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.
If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.