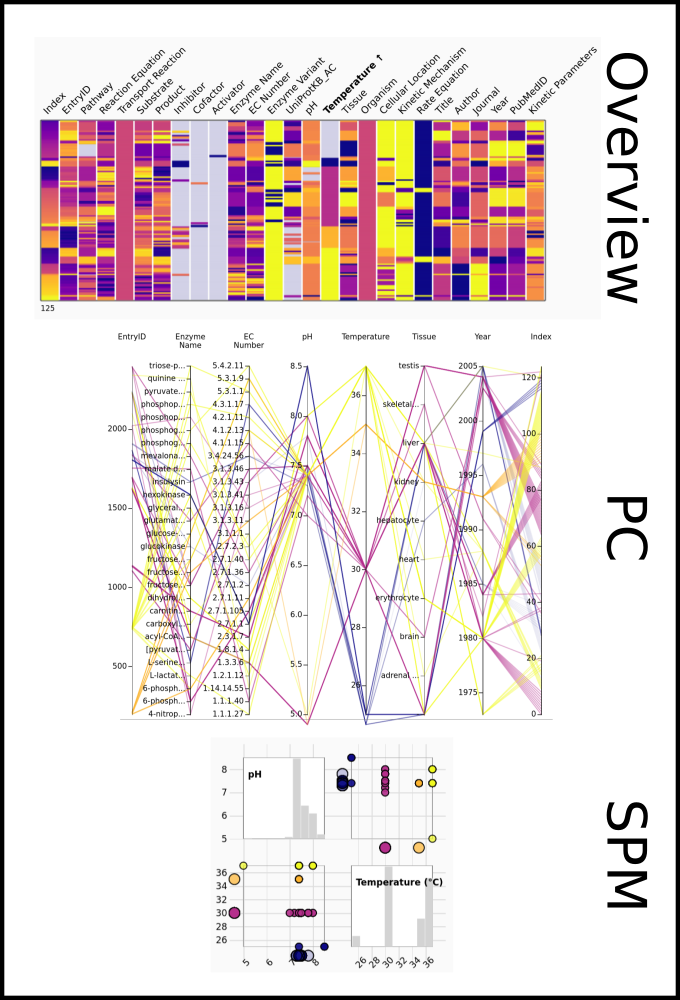
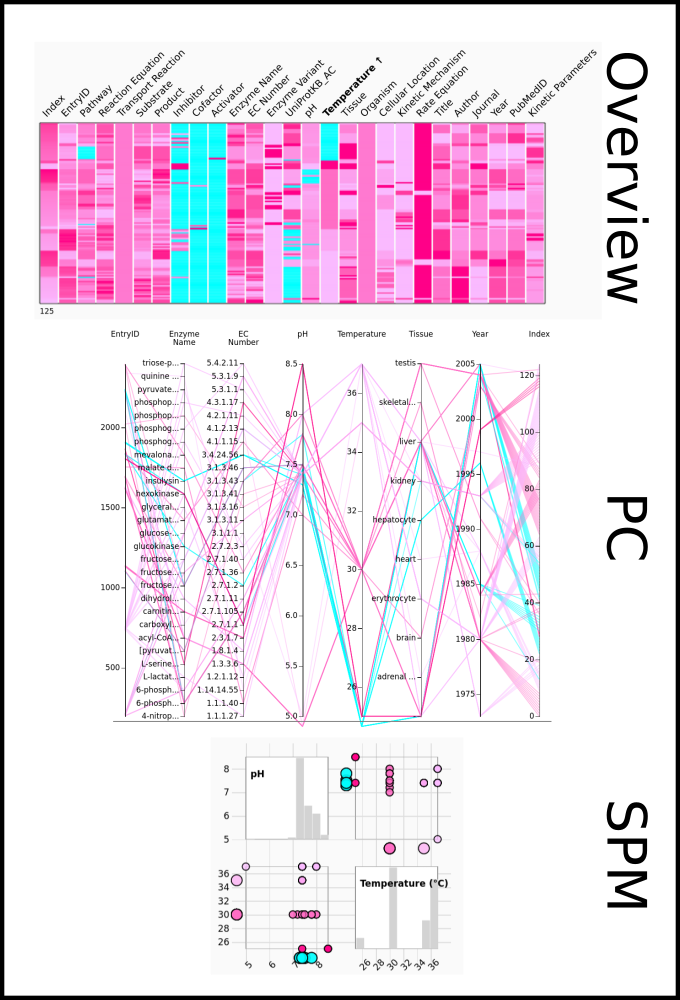
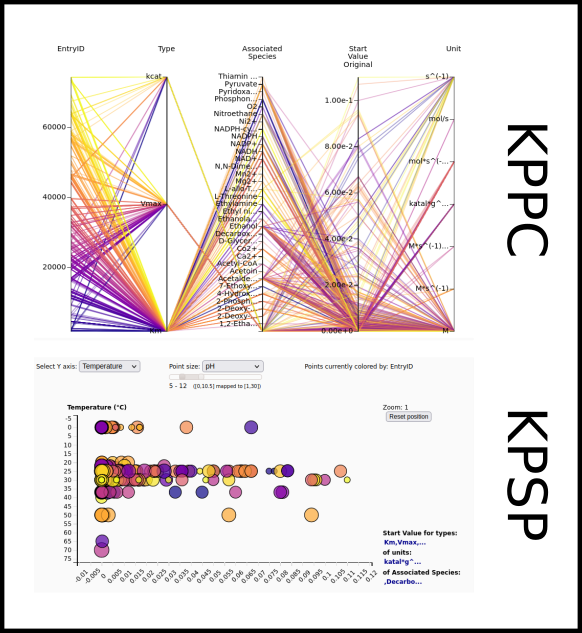
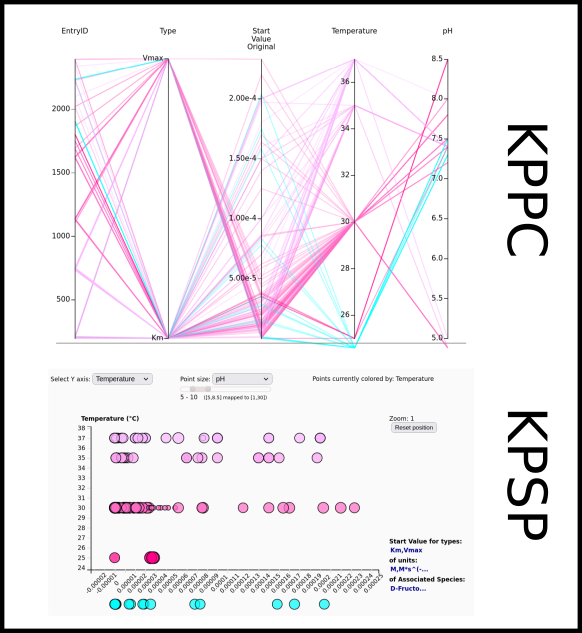
To improve SABIO-RK data interoperability, semantic markup was added to web pages as described and defined by
This structured information makes it easier to discover, collate and analyse our data.
|
32211
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32212
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32213
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32214
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32215
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32216
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32217
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32218
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32219
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32220
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32221
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32222
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32223
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32224
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32225
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32226
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32227
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32228
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32229
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32230
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32231
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32232
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32233
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32234
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32235
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32236
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32237
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32238
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32239
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32240
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32241
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32242
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32243
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32244
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32245
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32246
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32247
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32248
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32249
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32250
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32251
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32252
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32253
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32254
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32255
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32256
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32257
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32258
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32259
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32260
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32261
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32262
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32263
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32264
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32265
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32266
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32267
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32268
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32269
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32270
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32271
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32272
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32273
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32274
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32275
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32276
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32277
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32278
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32279
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32280
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32281
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32282
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32283
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32284
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32285
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32286
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32287
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32288
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32289
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32290
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32291
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32292
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32293
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32294
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32295
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32296
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32297
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32298
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32299
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32300
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32301
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32302
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32303
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32304
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32305
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32306
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32307
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32308
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32309
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32310
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32311
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32312
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32313
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32314
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32315
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32316
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32317
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32318
|
Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the ...
|
|
32319
|
Functional phosphoglucose isomerase from Mycobacterium tuberculosis H37Rv: rapid purification with high yield ...
|
|
32320
|
Functional phosphoglucose isomerase from Mycobacterium tuberculosis H37Rv: rapid purification with high yield ...
|
|
32321
|
Dynamic simulation of an in vitro multi-enzyme system
|
|
32322
|
Dynamic simulation of an in vitro multi-enzyme system
|
|
32323
|
Dynamic simulation of an in vitro multi-enzyme system
|
|
32324
|
Nucleotide sequence variation does not relate to differences in kinetic properties of neutral trehalase from ...
|
|
32325
|
Nucleotide sequence variation does not relate to differences in kinetic properties of neutral trehalase from ...
|
|
32326
|
Nucleotide sequence variation does not relate to differences in kinetic properties of neutral trehalase from ...
|
|
32327
|
Nucleotide sequence variation does not relate to differences in kinetic properties of neutral trehalase from ...
|
|
32328
|
Nucleotide sequence variation does not relate to differences in kinetic properties of neutral trehalase from ...
|
|
32329
|
Nucleotide sequence variation does not relate to differences in kinetic properties of neutral trehalase from ...
|
|
32330
|
Saccharomyces cerevisiae nucleoside-diphosphate kinase: purification, characterization, and substrate ...
|
|
32331
|
Saccharomyces cerevisiae nucleoside-diphosphate kinase: purification, characterization, and substrate ...
|
|
32332
|
Saccharomyces cerevisiae nucleoside-diphosphate kinase: purification, characterization, and substrate ...
|
|
32333
|
Saccharomyces cerevisiae nucleoside-diphosphate kinase: purification, characterization, and substrate ...
|
|
32334
|
Saccharomyces cerevisiae nucleoside-diphosphate kinase: purification, characterization, and substrate ...
|
|
32335
|
Saccharomyces cerevisiae nucleoside-diphosphate kinase: purification, characterization, and substrate ...
|
|
32336
|
Purification and properties of an extreme thermostable glutamate dehydrogenase from the archaebacterium ...
|
|
32337
|
Purification and properties of an extreme thermostable glutamate dehydrogenase from the archaebacterium ...
|
|
32338
|
Purification and properties of an extreme thermostable glutamate dehydrogenase from the archaebacterium ...
|
|
32339
|
Purification and properties of an extreme thermostable glutamate dehydrogenase from the archaebacterium ...
|
|
32340
|
Purification and properties of an extreme thermostable glutamate dehydrogenase from the archaebacterium ...
|
|
32341
|
Purification and properties of an extreme thermostable glutamate dehydrogenase from the archaebacterium ...
|
|
32342
|
Purification and properties of an extreme thermostable glutamate dehydrogenase from the archaebacterium ...
|
|
32343
|
Purification and properties of an extreme thermostable glutamate dehydrogenase from the archaebacterium ...
|
|
32344
|
Purification and properties of mitochondrial phosphoenolpyruvate carboxykinase from liver of Squalus acanthias
|
|
32345
|
Purification and properties of mitochondrial phosphoenolpyruvate carboxykinase from liver of Squalus acanthias
|
|
32346
|
Purification and properties of mitochondrial phosphoenolpyruvate carboxykinase from liver of Squalus acanthias
|
|
32347
|
Purification and properties of mitochondrial phosphoenolpyruvate carboxykinase from liver of Squalus acanthias
|
|
32348
|
Purification and properties of mitochondrial phosphoenolpyruvate carboxykinase from liver of Squalus acanthias
|
|
32349
|
Purification and properties of mitochondrial phosphoenolpyruvate carboxykinase from liver of Squalus acanthias
|
|
32350
|
Purification and properties of mitochondrial phosphoenolpyruvate carboxykinase from liver of Squalus acanthias
|
|
32351
|
Purification and properties of mitochondrial phosphoenolpyruvate carboxykinase from liver of Squalus acanthias
|
|
32352
|
Purification and properties of mitochondrial phosphoenolpyruvate carboxykinase from liver of Squalus acanthias
|
|
32353
|
Purification and properties of mitochondrial phosphoenolpyruvate carboxykinase from liver of Squalus acanthias
|
|
32354
|
Purification and properties of mitochondrial phosphoenolpyruvate carboxykinase from liver of Squalus acanthias
|
|
32355
|
Purification and properties of mitochondrial phosphoenolpyruvate carboxykinase from liver of Squalus acanthias
|
|
32356
|
Purification and properties of mitochondrial phosphoenolpyruvate carboxykinase from liver of Squalus acanthias
|
|
32362
|
Thermotoga maritima phosphofructokinases: expression and characterization of two unique enzymes
|
|
32363
|
Thermotoga maritima phosphofructokinases: expression and characterization of two unique enzymes
|
|
32364
|
Thermotoga maritima phosphofructokinases: expression and characterization of two unique enzymes
|
|
32365
|
Thermotoga maritima phosphofructokinases: expression and characterization of two unique enzymes
|
|
32366
|
Thermotoga maritima phosphofructokinases: expression and characterization of two unique enzymes
|
|
32367
|
Thermotoga maritima phosphofructokinases: expression and characterization of two unique enzymes
|
|
32368
|
Thermotoga maritima phosphofructokinases: expression and characterization of two unique enzymes
|
|
32369
|
Thermotoga maritima phosphofructokinases: expression and characterization of two unique enzymes
|
|
32370
|
Thermotoga maritima phosphofructokinases: expression and characterization of two unique enzymes
|
|
32371
|
Thermotoga maritima phosphofructokinases: expression and characterization of two unique enzymes
|
|
32372
|
Thermotoga maritima phosphofructokinases: expression and characterization of two unique enzymes
|
|
32373
|
Thermotoga maritima phosphofructokinases: expression and characterization of two unique enzymes
|
|
32383
|
H-tunneling in the multiple H-transfers of the catalytic cycle of morphinone reductase and in the reductive ...
|
|
32384
|
H-tunneling in the multiple H-transfers of the catalytic cycle of morphinone reductase and in the reductive ...
|
|
32385
|
H-tunneling in the multiple H-transfers of the catalytic cycle of morphinone reductase and in the reductive ...
|
|
32386
|
H-tunneling in the multiple H-transfers of the catalytic cycle of morphinone reductase and in the reductive ...
|
|
32387
|
H-tunneling in the multiple H-transfers of the catalytic cycle of morphinone reductase and in the reductive ...
|
|
32388
|
H-tunneling in the multiple H-transfers of the catalytic cycle of morphinone reductase and in the reductive ...
|
|
32389
|
H-tunneling in the multiple H-transfers of the catalytic cycle of morphinone reductase and in the reductive ...
|
|
32390
|
Control of glycolytic dynamics by hexose transport in Saccharomyces cerevisiae
|
|
32391
|
Control of glycolytic dynamics by hexose transport in Saccharomyces cerevisiae
|
|
32392
|
Control of glycolytic dynamics by hexose transport in Saccharomyces cerevisiae
|
|
32393
|
Control of glycolytic dynamics by hexose transport in Saccharomyces cerevisiae
|
|
32394
|
Microsomal 17beta-hydroxysteroid dehydrogenase of guinea pig liver: detergent solubilization and a comparison ...
|
|
32395
|
Microsomal 17beta-hydroxysteroid dehydrogenase of guinea pig liver: detergent solubilization and a comparison ...
|
|
32396
|
Microsomal 17beta-hydroxysteroid dehydrogenase of guinea pig liver: detergent solubilization and a comparison ...
|
|
32397
|
Microsomal 17beta-hydroxysteroid dehydrogenase of guinea pig liver: detergent solubilization and a comparison ...
|
|
32398
|
Reduction of mevaldic acid to mevalonic acid by a partially purified enzyme from liver
|
|
32399
|
Reduction of mevaldic acid to mevalonic acid by a partially purified enzyme from liver
|
|
32400
|
Reduction of mevaldic acid to mevalonic acid by a partially purified enzyme from liver
|
|
32401
|
Functional characterization of wild-type and variant (T202I and M59I) human UDP-glucuronosyltransferase 1A10
|
|
32402
|
Functional characterization of wild-type and variant (T202I and M59I) human UDP-glucuronosyltransferase 1A10
|
|
32403
|
Functional characterization of wild-type and variant (T202I and M59I) human UDP-glucuronosyltransferase 1A10
|
|
32404
|
Functional characterization of wild-type and variant (T202I and M59I) human UDP-glucuronosyltransferase 1A10
|
|
32405
|
Functional characterization of wild-type and variant (T202I and M59I) human UDP-glucuronosyltransferase 1A10
|
|
32406
|
Functional characterization of wild-type and variant (T202I and M59I) human UDP-glucuronosyltransferase 1A10
|
|
32407
|
Characterization of an extremely thermostable glutamate dehydrogenase: a key enzyme in the primary metabolism ...
|
|
32408
|
Characterization of an extremely thermostable glutamate dehydrogenase: a key enzyme in the primary metabolism ...
|
|
32409
|
Characterization of an extremely thermostable glutamate dehydrogenase: a key enzyme in the primary metabolism ...
|
|
32410
|
Characterization of an extremely thermostable glutamate dehydrogenase: a key enzyme in the primary metabolism ...
|
|
32411
|
Characterization of an extremely thermostable glutamate dehydrogenase: a key enzyme in the primary metabolism ...
|
|
32412
|
Characterization of an extremely thermostable glutamate dehydrogenase: a key enzyme in the primary metabolism ...
|
|
32413
|
Characterization of an extremely thermostable glutamate dehydrogenase: a key enzyme in the primary metabolism ...
|
|
32414
|
Characterization of an extremely thermostable glutamate dehydrogenase: a key enzyme in the primary metabolism ...
|
|
32415
|
Characterization of an extremely thermostable glutamate dehydrogenase: a key enzyme in the primary metabolism ...
|
|
32416
|
Characterization and regulatory properties of a single hexokinase from the citric acid accumulating fungus ...
|
|
32417
|
Characterization and regulatory properties of a single hexokinase from the citric acid accumulating fungus ...
|
|
32418
|
Characterization and regulatory properties of a single hexokinase from the citric acid accumulating fungus ...
|
|
32419
|
Characterization and regulatory properties of a single hexokinase from the citric acid accumulating fungus ...
|
|
32420
|
Phosphofructokinase from white muscle of the rainbow trout, Oncorhynchus mykiss: purification and properties
|
|
32421
|
Phosphofructokinase from white muscle of the rainbow trout, Oncorhynchus mykiss: purification and properties
|
|
32422
|
Phosphofructokinase from white muscle of the rainbow trout, Oncorhynchus mykiss: purification and properties
|
|
32423
|
Phosphofructokinase from white muscle of the rainbow trout, Oncorhynchus mykiss: purification and properties
|
|
32424
|
Phosphofructokinase from white muscle of the rainbow trout, Oncorhynchus mykiss: purification and properties
|
|
32425
|
Phosphofructokinase from white muscle of the rainbow trout, Oncorhynchus mykiss: purification and properties
|
|
32426
|
Phosphofructokinase from white muscle of the rainbow trout, Oncorhynchus mykiss: purification and properties
|
|
32427
|
Phosphofructokinase from white muscle of the rainbow trout, Oncorhynchus mykiss: purification and properties
|
|
32428
|
Purification and characterization of glutamate dehydrogenase as another isoprotein binding to the membrane of ...
|
|
32429
|
Purification and characterization of glutamate dehydrogenase as another isoprotein binding to the membrane of ...
|
|
32430
|
Purification and characterization of glutamate dehydrogenase as another isoprotein binding to the membrane of ...
|
|
32431
|
Purification and characterization of glutamate dehydrogenase as another isoprotein binding to the membrane of ...
|
|
32432
|
Purification and characterization of glutamate dehydrogenase as another isoprotein binding to the membrane of ...
|
|
32433
|
Purification and characterization of glutamate dehydrogenase as another isoprotein binding to the membrane of ...
|
|
32434
|
Purification and characterization of glutamate dehydrogenase as another isoprotein binding to the membrane of ...
|
|
32435
|
Purification and characterization of glutamate dehydrogenase as another isoprotein binding to the membrane of ...
|
|
32436
|
Identification and characterization of single-domain thiosulfate sulfurtransferases from Arabidopsis thaliana
|
|
32437
|
Identification and characterization of single-domain thiosulfate sulfurtransferases from Arabidopsis thaliana
|
|
32438
|
Identification and characterization of single-domain thiosulfate sulfurtransferases from Arabidopsis thaliana
|
|
32439
|
Identification and characterization of single-domain thiosulfate sulfurtransferases from Arabidopsis thaliana
|
|
32440
|
Identification and characterization of single-domain thiosulfate sulfurtransferases from Arabidopsis thaliana
|
|
32441
|
Identification and characterization of single-domain thiosulfate sulfurtransferases from Arabidopsis thaliana
|
|
32442
|
Importance of glutamate 279 for the coenzyme binding of human glutamate dehydrogenase
|
|
32443
|
Importance of glutamate 279 for the coenzyme binding of human glutamate dehydrogenase
|
|
32444
|
Importance of glutamate 279 for the coenzyme binding of human glutamate dehydrogenase
|
|
32445
|
Importance of glutamate 279 for the coenzyme binding of human glutamate dehydrogenase
|
|
32446
|
Importance of glutamate 279 for the coenzyme binding of human glutamate dehydrogenase
|
|
32447
|
Importance of glutamate 279 for the coenzyme binding of human glutamate dehydrogenase
|
|
32448
|
Importance of glutamate 279 for the coenzyme binding of human glutamate dehydrogenase
|
|
32449
|
Importance of glutamate 279 for the coenzyme binding of human glutamate dehydrogenase
|
|
32450
|
Importance of glutamate 279 for the coenzyme binding of human glutamate dehydrogenase
|
|
32451
|
Importance of glutamate 279 for the coenzyme binding of human glutamate dehydrogenase
|
|
32452
|
Importance of glutamate 279 for the coenzyme binding of human glutamate dehydrogenase
|
|
32453
|
Importance of glutamate 279 for the coenzyme binding of human glutamate dehydrogenase
|
|
32454
|
Isolation and partial purification of mitochondrial and cytosolic rhodanese from liver of normal and ...
|
|
32455
|
Isolation and partial purification of mitochondrial and cytosolic rhodanese from liver of normal and ...
|
|
32456
|
Isolation and partial purification of mitochondrial and cytosolic rhodanese from liver of normal and ...
|
|
32457
|
Isolation and partial purification of mitochondrial and cytosolic rhodanese from liver of normal and ...
|
|
32458
|
Isolation and partial purification of mitochondrial and cytosolic rhodanese from liver of normal and ...
|
|
32459
|
Isolation and partial purification of mitochondrial and cytosolic rhodanese from liver of normal and ...
|
|
32460
|
Isolation and partial purification of mitochondrial and cytosolic rhodanese from liver of normal and ...
|
|
32461
|
Isolation and partial purification of mitochondrial and cytosolic rhodanese from liver of normal and ...
|
|
32462
|
Cold adaptation of xylose isomerase from Thermus thermophilus through random PCR mutagenesis. Gene cloning and ...
|
|
32463
|
Cold adaptation of xylose isomerase from Thermus thermophilus through random PCR mutagenesis. Gene cloning and ...
|
|
32464
|
Cold adaptation of xylose isomerase from Thermus thermophilus through random PCR mutagenesis. Gene cloning and ...
|
|
32465
|
Cold adaptation of xylose isomerase from Thermus thermophilus through random PCR mutagenesis. Gene cloning and ...
|
|
32466
|
Cold adaptation of xylose isomerase from Thermus thermophilus through random PCR mutagenesis. Gene cloning and ...
|
|
32467
|
Cold adaptation of xylose isomerase from Thermus thermophilus through random PCR mutagenesis. Gene cloning and ...
|
|
32468
|
Cold adaptation of xylose isomerase from Thermus thermophilus through random PCR mutagenesis. Gene cloning and ...
|
|
32469
|
Cold adaptation of xylose isomerase from Thermus thermophilus through random PCR mutagenesis. Gene cloning and ...
|
|
32470
|
Cold adaptation of xylose isomerase from Thermus thermophilus through random PCR mutagenesis. Gene cloning and ...
|
|
32471
|
Cold adaptation of xylose isomerase from Thermus thermophilus through random PCR mutagenesis. Gene cloning and ...
|
|
32472
|
Cold adaptation of xylose isomerase from Thermus thermophilus through random PCR mutagenesis. Gene cloning and ...
|
|
32473
|
Cold adaptation of xylose isomerase from Thermus thermophilus through random PCR mutagenesis. Gene cloning and ...
|
|
32474
|
Study of the mode of action of endopolygalacturonase from Fusarium moniliforme
|
|
32475
|
Study of the mode of action of endopolygalacturonase from Fusarium moniliforme
|
|
32476
|
Study of the mode of action of endopolygalacturonase from Fusarium moniliforme
|
|
32477
|
Study of the mode of action of endopolygalacturonase from Fusarium moniliforme
|
|
32478
|
Study of the mode of action of endopolygalacturonase from Fusarium moniliforme
|
|
32479
|
Study of the mode of action of endopolygalacturonase from Fusarium moniliforme
|
|
32480
|
Partial purification and characterization of L-lactate dehydrogenase isozymes from sweet potato roots
|
|
32481
|
Partial purification and characterization of L-lactate dehydrogenase isozymes from sweet potato roots
|
|
32482
|
Partial purification and characterization of L-lactate dehydrogenase isozymes from sweet potato roots
|
|
32483
|
Partial purification and characterization of L-lactate dehydrogenase isozymes from sweet potato roots
|
|
32484
|
Partial purification and characterization of L-lactate dehydrogenase isozymes from sweet potato roots
|
|
32485
|
Partial purification and characterization of L-lactate dehydrogenase isozymes from sweet potato roots
|
|
32486
|
Partial purification and characterization of L-lactate dehydrogenase isozymes from sweet potato roots
|
|
32487
|
Partial purification and characterization of L-lactate dehydrogenase isozymes from sweet potato roots
|
|
32488
|
Partial purification and characterization of L-lactate dehydrogenase isozymes from sweet potato roots
|
|
32489
|
Partial purification and characterization of L-lactate dehydrogenase isozymes from sweet potato roots
|
|
32490
|
Competitive inhibition of liver glucokinase by its regulatory protein
|
|
32491
|
Competitive inhibition of liver glucokinase by its regulatory protein
|
|
32492
|
Competitive inhibition of liver glucokinase by its regulatory protein
|
|
32493
|
Competitive inhibition of liver glucokinase by its regulatory protein
|
|
32494
|
Competitive inhibition of liver glucokinase by its regulatory protein
|
|
32495
|
Competitive inhibition of liver glucokinase by its regulatory protein
|
|
32496
|
Competitive inhibition of liver glucokinase by its regulatory protein
|
|
32497
|
Competitive inhibition of liver glucokinase by its regulatory protein
|
|
32498
|
Competitive inhibition of liver glucokinase by its regulatory protein
|
|
32499
|
Competitive inhibition of liver glucokinase by its regulatory protein
|
|
32500
|
Competitive inhibition of liver glucokinase by its regulatory protein
|
|
32501
|
Competitive inhibition of liver glucokinase by its regulatory protein
|
|
32502
|
Competitive inhibition of liver glucokinase by its regulatory protein
|
|
32503
|
Competitive inhibition of liver glucokinase by its regulatory protein
|
|
32504
|
Substrate specificity and structure-activity relationships of gentamicin acetyltransferase I. The dependence ...
|
|
32505
|
Substrate specificity and structure-activity relationships of gentamicin acetyltransferase I. The dependence ...
|
|
32506
|
Substrate specificity and structure-activity relationships of gentamicin acetyltransferase I. The dependence ...
|
|
32507
|
Substrate specificity and structure-activity relationships of gentamicin acetyltransferase I. The dependence ...
|
|
32508
|
Substrate specificity and structure-activity relationships of gentamicin acetyltransferase I. The dependence ...
|
|
32509
|
Substrate specificity and structure-activity relationships of gentamicin acetyltransferase I. The dependence ...
|
|
32510
|
Substrate specificity and structure-activity relationships of gentamicin acetyltransferase I. The dependence ...
|
|
32511
|
Substrate specificity and structure-activity relationships of gentamicin acetyltransferase I. The dependence ...
|
|
32512
|
Substrate specificity and structure-activity relationships of gentamicin acetyltransferase I. The dependence ...
|
|
32513
|
Substrate specificity and structure-activity relationships of gentamicin acetyltransferase I. The dependence ...
|
|
32514
|
Substrate specificity and structure-activity relationships of gentamicin acetyltransferase I. The dependence ...
|
|
32515
|
Substrate specificity and structure-activity relationships of gentamicin acetyltransferase I. The dependence ...
|
|
32516
|
Substrate specificity and structure-activity relationships of gentamicin acetyltransferase I. The dependence ...
|
|
32517
|
Substrate specificity and structure-activity relationships of gentamicin acetyltransferase I. The dependence ...
|
|
32518
|
Substrate specificity and structure-activity relationships of gentamicin acetyltransferase I. The dependence ...
|
|
32519
|
Substrate specificity and structure-activity relationships of gentamicin acetyltransferase I. The dependence ...
|
|
32520
|
Substrate specificity and structure-activity relationships of gentamicin acetyltransferase I. The dependence ...
|
|
32521
|
Substrate specificity and structure-activity relationships of gentamicin acetyltransferase I. The dependence ...
|
|
32522
|
Substrate specificity and structure-activity relationships of gentamicin acetyltransferase I. The dependence ...
|
|
32523
|
Substrate specificity and structure-activity relationships of gentamicin acetyltransferase I. The dependence ...
|
|
32524
|
Substrate specificity and structure-activity relationships of gentamicin acetyltransferase I. The dependence ...
|
|
32525
|
Substrate specificity and structure-activity relationships of gentamicin acetyltransferase I. The dependence ...
|
|
32526
|
Substrate specificity and structure-activity relationships of gentamicin acetyltransferase I. The dependence ...
|
|
32527
|
Substrate specificity and structure-activity relationships of gentamicin acetyltransferase I. The dependence ...
|
|
32528
|
Substrate specificity and structure-activity relationships of gentamicin acetyltransferase I. The dependence ...
|
|
32529
|
Inhibition of phosphoglycerate kinase by products and product homologues
|
|
32530
|
Inhibition of phosphoglycerate kinase by products and product homologues
|
|
32531
|
Inhibition of phosphoglycerate kinase by products and product homologues
|
|
32532
|
Inhibition of phosphoglycerate kinase by products and product homologues
|
|
32533
|
Inhibition of phosphoglycerate kinase by products and product homologues
|
|
32534
|
Inhibition of phosphoglycerate kinase by products and product homologues
|
|
32535
|
Inhibition of phosphoglycerate kinase by products and product homologues
|
|
32536
|
Inhibition of phosphoglycerate kinase by products and product homologues
|
|
32537
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32538
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32539
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32540
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32541
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32542
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32543
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32544
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32545
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32546
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32547
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32548
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32549
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32550
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32551
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32552
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32553
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32554
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32555
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32556
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32557
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32558
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32559
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32560
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32561
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32562
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32563
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32564
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32565
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32566
|
The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from ...
|
|
32567
|
Trypanosoma brucei contains a 2,3-bisphosphoglycerate independent phosphoglycerate mutase
|
|
32568
|
Trypanosoma brucei contains a 2,3-bisphosphoglycerate independent phosphoglycerate mutase
|
|
32569
|
Purification and characterisation of an alpha-glucan phosphorylase from the thermophilic bacterium Thermus ...
|
|
32570
|
Purification and characterisation of an alpha-glucan phosphorylase from the thermophilic bacterium Thermus ...
|
|
32571
|
Purification and characterisation of an alpha-glucan phosphorylase from the thermophilic bacterium Thermus ...
|
|
32572
|
Purification and characterisation of an alpha-glucan phosphorylase from the thermophilic bacterium Thermus ...
|
|
32573
|
Purification and characterisation of an alpha-glucan phosphorylase from the thermophilic bacterium Thermus ...
|
|
32574
|
Purification and characterisation of an alpha-glucan phosphorylase from the thermophilic bacterium Thermus ...
|
|
32575
|
Purification and characterisation of an alpha-glucan phosphorylase from the thermophilic bacterium Thermus ...
|
|
32576
|
Purification and characterisation of an alpha-glucan phosphorylase from the thermophilic bacterium Thermus ...
|
|
32577
|
Purification and characterisation of an alpha-glucan phosphorylase from the thermophilic bacterium Thermus ...
|
|
32578
|
Purification and characterisation of an alpha-glucan phosphorylase from the thermophilic bacterium Thermus ...
|
|
32579
|
Purification and characterisation of an alpha-glucan phosphorylase from the thermophilic bacterium Thermus ...
|
|
32580
|
Purification and characterisation of an alpha-glucan phosphorylase from the thermophilic bacterium Thermus ...
|
|
32581
|
Purification and characterisation of an alpha-glucan phosphorylase from the thermophilic bacterium Thermus ...
|
|
32582
|
Purification and characterisation of an alpha-glucan phosphorylase from the thermophilic bacterium Thermus ...
|
|
32583
|
Purification and characterisation of an alpha-glucan phosphorylase from the thermophilic bacterium Thermus ...
|
|
32584
|
Purification and characterisation of an alpha-glucan phosphorylase from the thermophilic bacterium Thermus ...
|
|
32585
|
Comparative study of Leishmania mexicana and Trypanosoma brucei NAD-dependent glycerol-3-phosphate ...
|
|
32586
|
Comparative study of Leishmania mexicana and Trypanosoma brucei NAD-dependent glycerol-3-phosphate ...
|
|
32587
|
Comparative study of Leishmania mexicana and Trypanosoma brucei NAD-dependent glycerol-3-phosphate ...
|
|
32588
|
Comparative study of Leishmania mexicana and Trypanosoma brucei NAD-dependent glycerol-3-phosphate ...
|
|
32589
|
Comparative study of Leishmania mexicana and Trypanosoma brucei NAD-dependent glycerol-3-phosphate ...
|
|
32590
|
Comparative study of Leishmania mexicana and Trypanosoma brucei NAD-dependent glycerol-3-phosphate ...
|
|
32591
|
Comparative study of Leishmania mexicana and Trypanosoma brucei NAD-dependent glycerol-3-phosphate ...
|
|
32592
|
Comparative study of Leishmania mexicana and Trypanosoma brucei NAD-dependent glycerol-3-phosphate ...
|
|
32593
|
Comparative study of Leishmania mexicana and Trypanosoma brucei NAD-dependent glycerol-3-phosphate ...
|
|
32594
|
Comparative study of Leishmania mexicana and Trypanosoma brucei NAD-dependent glycerol-3-phosphate ...
|
|
32595
|
Comparative study of Leishmania mexicana and Trypanosoma brucei NAD-dependent glycerol-3-phosphate ...
|
|
32596
|
Comparative study of Leishmania mexicana and Trypanosoma brucei NAD-dependent glycerol-3-phosphate ...
|
|
32597
|
Comparative study of Leishmania mexicana and Trypanosoma brucei NAD-dependent glycerol-3-phosphate ...
|
|
32598
|
Comparative study of Leishmania mexicana and Trypanosoma brucei NAD-dependent glycerol-3-phosphate ...
|
|
32599
|
Comparative study of Leishmania mexicana and Trypanosoma brucei NAD-dependent glycerol-3-phosphate ...
|
|
32600
|
Cassette mutagenesis and photoaffinity labeling of adenine binding domain of ADP regulatory site within human ...
|
|
32601
|
Cassette mutagenesis and photoaffinity labeling of adenine binding domain of ADP regulatory site within human ...
|
|
32602
|
Cassette mutagenesis and photoaffinity labeling of adenine binding domain of ADP regulatory site within human ...
|
|
32603
|
Cassette mutagenesis and photoaffinity labeling of adenine binding domain of ADP regulatory site within human ...
|
|
32604
|
Cassette mutagenesis and photoaffinity labeling of adenine binding domain of ADP regulatory site within human ...
|
|
32605
|
Cassette mutagenesis and photoaffinity labeling of adenine binding domain of ADP regulatory site within human ...
|
|
32606
|
Cassette mutagenesis and photoaffinity labeling of adenine binding domain of ADP regulatory site within human ...
|
|
32607
|
Cassette mutagenesis and photoaffinity labeling of adenine binding domain of ADP regulatory site within human ...
|
|
32608
|
Cassette mutagenesis and photoaffinity labeling of adenine binding domain of ADP regulatory site within human ...
|
|
32609
|
Cassette mutagenesis and photoaffinity labeling of adenine binding domain of ADP regulatory site within human ...
|
|
32610
|
Cassette mutagenesis and photoaffinity labeling of adenine binding domain of ADP regulatory site within human ...
|
|
32611
|
Cassette mutagenesis and photoaffinity labeling of adenine binding domain of ADP regulatory site within human ...
|
|
32612
|
Purification and characterization of a thermostable class II fumarase from Thermus thermophilus
|
|
32613
|
Purification and characterization of a thermostable class II fumarase from Thermus thermophilus
|
|
32614
|
Purification and characterization of a thermostable class II fumarase from Thermus thermophilus
|
|
32615
|
Purification and characterization of a thermostable class II fumarase from Thermus thermophilus
|
|
32616
|
Purification and characterization of a thermostable class II fumarase from Thermus thermophilus
|
|
32617
|
Purification and cloning of a thermostable xylose (glucose) isomerase with an acidic pH optimum from ...
|
|
32618
|
Purification and cloning of a thermostable xylose (glucose) isomerase with an acidic pH optimum from ...
|
|
32619
|
Purification and cloning of a thermostable xylose (glucose) isomerase with an acidic pH optimum from ...
|
|
32620
|
Copy-DNA cloning and characterisation of a potato alpha-glucosidase: expression in Escherichia coli and ...
|
|
32621
|
Copy-DNA cloning and characterisation of a potato alpha-glucosidase: expression in Escherichia coli and ...
|
|
32622
|
Copy-DNA cloning and characterisation of a potato alpha-glucosidase: expression in Escherichia coli and ...
|
|
32623
|
Copy-DNA cloning and characterisation of a potato alpha-glucosidase: expression in Escherichia coli and ...
|
|
32624
|
Copy-DNA cloning and characterisation of a potato alpha-glucosidase: expression in Escherichia coli and ...
|
|
32625
|
Copy-DNA cloning and characterisation of a potato alpha-glucosidase: expression in Escherichia coli and ...
|
|
32626
|
Copy-DNA cloning and characterisation of a potato alpha-glucosidase: expression in Escherichia coli and ...
|
|
32627
|
Biochemical characterization of recombinant phosphoglucose isomerase of Mycobacterium tuberculosis
|
|
32628
|
Biochemical characterization of recombinant phosphoglucose isomerase of Mycobacterium tuberculosis
|
|
32629
|
Purification, properties, and evidence for two subtypes of human placenta hexokinase type I
|
|
32630
|
Purification, properties, and evidence for two subtypes of human placenta hexokinase type I
|
|
32631
|
Purification, properties, and evidence for two subtypes of human placenta hexokinase type I
|
|
32632
|
Purification, properties, and evidence for two subtypes of human placenta hexokinase type I
|
|
32633
|
Purification, properties, and evidence for two subtypes of human placenta hexokinase type I
|
|
32634
|
Purification, properties, and evidence for two subtypes of human placenta hexokinase type I
|
|
32635
|
Purification, properties, and evidence for two subtypes of human placenta hexokinase type I
|
|
32636
|
Purification, properties, and evidence for two subtypes of human placenta hexokinase type I
|
|
32637
|
Purification, properties, and evidence for two subtypes of human placenta hexokinase type I
|
|
32638
|
Inhibition of hexonate dehydrogenase and aldose reductase from bovine retina by sorbinil, statil, M79175 and ...
|
|
32639
|
Inhibition of hexonate dehydrogenase and aldose reductase from bovine retina by sorbinil, statil, M79175 and ...
|
|
32640
|
Inhibition of hexonate dehydrogenase and aldose reductase from bovine retina by sorbinil, statil, M79175 and ...
|
|
32641
|
Inhibition of hexonate dehydrogenase and aldose reductase from bovine retina by sorbinil, statil, M79175 and ...
|
|
32642
|
Inhibition of hexonate dehydrogenase and aldose reductase from bovine retina by sorbinil, statil, M79175 and ...
|
|
32643
|
Inhibition of hexonate dehydrogenase and aldose reductase from bovine retina by sorbinil, statil, M79175 and ...
|
|
32644
|
Inhibition of hexonate dehydrogenase and aldose reductase from bovine retina by sorbinil, statil, M79175 and ...
|
|
32645
|
Inhibition of hexonate dehydrogenase and aldose reductase from bovine retina by sorbinil, statil, M79175 and ...
|
|
32646
|
Inhibition of hexonate dehydrogenase and aldose reductase from bovine retina by sorbinil, statil, M79175 and ...
|
|
32647
|
Inhibition of hexonate dehydrogenase and aldose reductase from bovine retina by sorbinil, statil, M79175 and ...
|
|
32648
|
Inhibition of hexonate dehydrogenase and aldose reductase from bovine retina by sorbinil, statil, M79175 and ...
|
|
32649
|
Inhibition of hexonate dehydrogenase and aldose reductase from bovine retina by sorbinil, statil, M79175 and ...
|
|
32650
|
Examination of the thiamin diphosphate binding site in yeast transketolase by site-directed mutagenesis
|
|
32651
|
Examination of the thiamin diphosphate binding site in yeast transketolase by site-directed mutagenesis
|
|
32652
|
Examination of the thiamin diphosphate binding site in yeast transketolase by site-directed mutagenesis
|
|
32653
|
Examination of the thiamin diphosphate binding site in yeast transketolase by site-directed mutagenesis
|
|
32654
|
Examination of the thiamin diphosphate binding site in yeast transketolase by site-directed mutagenesis
|
|
32655
|
Examination of the thiamin diphosphate binding site in yeast transketolase by site-directed mutagenesis
|
|
32656
|
Examination of the thiamin diphosphate binding site in yeast transketolase by site-directed mutagenesis
|
|
32657
|
Examination of the thiamin diphosphate binding site in yeast transketolase by site-directed mutagenesis
|
|
32658
|
Examination of the thiamin diphosphate binding site in yeast transketolase by site-directed mutagenesis
|
|
32659
|
Examination of the thiamin diphosphate binding site in yeast transketolase by site-directed mutagenesis
|
|
32660
|
Examination of the thiamin diphosphate binding site in yeast transketolase by site-directed mutagenesis
|
|
32661
|
Examination of the thiamin diphosphate binding site in yeast transketolase by site-directed mutagenesis
|
|
32662
|
Examination of the thiamin diphosphate binding site in yeast transketolase by site-directed mutagenesis
|
|
32663
|
Examination of the thiamin diphosphate binding site in yeast transketolase by site-directed mutagenesis
|
|
32664
|
Examination of the thiamin diphosphate binding site in yeast transketolase by site-directed mutagenesis
|
|
32665
|
Human aldehyde dehydrogenase: kinetic identification of the isozyme for which biogenic aldehydes and ...
|
|
32666
|
Human aldehyde dehydrogenase: kinetic identification of the isozyme for which biogenic aldehydes and ...
|
|
32667
|
Human aldehyde dehydrogenase: kinetic identification of the isozyme for which biogenic aldehydes and ...
|
|
32668
|
Human aldehyde dehydrogenase: kinetic identification of the isozyme for which biogenic aldehydes and ...
|
|
32669
|
Human aldehyde dehydrogenase: kinetic identification of the isozyme for which biogenic aldehydes and ...
|
|
32670
|
Human aldehyde dehydrogenase: kinetic identification of the isozyme for which biogenic aldehydes and ...
|
|
32671
|
Human aldehyde dehydrogenase: kinetic identification of the isozyme for which biogenic aldehydes and ...
|
|
32672
|
Human aldehyde dehydrogenase: kinetic identification of the isozyme for which biogenic aldehydes and ...
|
|
32673
|
Human aldehyde dehydrogenase: kinetic identification of the isozyme for which biogenic aldehydes and ...
|
|
32674
|
Human aldehyde dehydrogenase: kinetic identification of the isozyme for which biogenic aldehydes and ...
|
|
32675
|
Human aldehyde dehydrogenase: kinetic identification of the isozyme for which biogenic aldehydes and ...
|
|
32676
|
Human aldehyde dehydrogenase: kinetic identification of the isozyme for which biogenic aldehydes and ...
|
|
32677
|
Overproduction, purification, and characterization of recombinant bifunctional threonine-sensitive aspartate ...
|
|
32678
|
Overproduction, purification, and characterization of recombinant bifunctional threonine-sensitive aspartate ...
|
|
32679
|
Overproduction, purification, and characterization of recombinant bifunctional threonine-sensitive aspartate ...
|
|
32680
|
Overproduction, purification, and characterization of recombinant bifunctional threonine-sensitive aspartate ...
|
|
32681
|
Overproduction, purification, and characterization of recombinant bifunctional threonine-sensitive aspartate ...
|
|
32682
|
Overproduction, purification, and characterization of recombinant bifunctional threonine-sensitive aspartate ...
|
|
32683
|
Overproduction, purification, and characterization of recombinant bifunctional threonine-sensitive aspartate ...
|
|
32684
|
Molecular cloning and characterization of Brugia malayi hexokinase
|
|
32685
|
Molecular cloning and characterization of Brugia malayi hexokinase
|
|
32686
|
Molecular cloning and characterization of Brugia malayi hexokinase
|
|
32687
|
Molecular cloning and characterization of Brugia malayi hexokinase
|
|
32688
|
Molecular cloning and characterization of Brugia malayi hexokinase
|
|
32689
|
Different physiological roles of ATP- and PP(i)-dependent phosphofructokinase isoenzymes in the methylotrophic ...
|
|
32690
|
Different physiological roles of ATP- and PP(i)-dependent phosphofructokinase isoenzymes in the methylotrophic ...
|
|
32691
|
Different physiological roles of ATP- and PP(i)-dependent phosphofructokinase isoenzymes in the methylotrophic ...
|
|
32692
|
Different physiological roles of ATP- and PP(i)-dependent phosphofructokinase isoenzymes in the methylotrophic ...
|
|
32693
|
Different physiological roles of ATP- and PP(i)-dependent phosphofructokinase isoenzymes in the methylotrophic ...
|
|
32694
|
Different physiological roles of ATP- and PP(i)-dependent phosphofructokinase isoenzymes in the methylotrophic ...
|
|
32695
|
Different physiological roles of ATP- and PP(i)-dependent phosphofructokinase isoenzymes in the methylotrophic ...
|
|
32696
|
Different physiological roles of ATP- and PP(i)-dependent phosphofructokinase isoenzymes in the methylotrophic ...
|
|
32697
|
Sequence analysis of glutamate dehydrogenase (GDH) from the hyperthermophilic archaeon Pyrococcus sp. KOD1 and ...
|
|
32698
|
Sequence analysis of glutamate dehydrogenase (GDH) from the hyperthermophilic archaeon Pyrococcus sp. KOD1 and ...
|
|
32699
|
Sequence analysis of glutamate dehydrogenase (GDH) from the hyperthermophilic archaeon Pyrococcus sp. KOD1 and ...
|
|
32700
|
Sequence analysis of glutamate dehydrogenase (GDH) from the hyperthermophilic archaeon Pyrococcus sp. KOD1 and ...
|
|
32701
|
Sequence analysis of glutamate dehydrogenase (GDH) from the hyperthermophilic archaeon Pyrococcus sp. KOD1 and ...
|
|
32702
|
Sequence analysis of glutamate dehydrogenase (GDH) from the hyperthermophilic archaeon Pyrococcus sp. KOD1 and ...
|
|
32703
|
Sequence analysis of glutamate dehydrogenase (GDH) from the hyperthermophilic archaeon Pyrococcus sp. KOD1 and ...
|
|
32704
|
Sequence analysis of glutamate dehydrogenase (GDH) from the hyperthermophilic archaeon Pyrococcus sp. KOD1 and ...
|
|
32705
|
Studies on the substrate specificity of Escherichia coli galactokinase
|
|
32706
|
Studies on the substrate specificity of Escherichia coli galactokinase
|
|
32707
|
Studies on the substrate specificity of Escherichia coli galactokinase
|
|
32708
|
Studies on the substrate specificity of Escherichia coli galactokinase
|
|
32709
|
Studies on the substrate specificity of Escherichia coli galactokinase
|
|
32710
|
Studies on the substrate specificity of Escherichia coli galactokinase
|
|
32711
|
Islet fructose 6-phosphate, 2-kinase:fructose 2,6-bisphosphatase: isozymic form, expression, and ...
|
|
32712
|
Islet fructose 6-phosphate, 2-kinase:fructose 2,6-bisphosphatase: isozymic form, expression, and ...
|
|
32713
|
Islet fructose 6-phosphate, 2-kinase:fructose 2,6-bisphosphatase: isozymic form, expression, and ...
|
|
32714
|
Islet fructose 6-phosphate, 2-kinase:fructose 2,6-bisphosphatase: isozymic form, expression, and ...
|
|
32715
|
Islet fructose 6-phosphate, 2-kinase:fructose 2,6-bisphosphatase: isozymic form, expression, and ...
|
|
32716
|
Islet fructose 6-phosphate, 2-kinase:fructose 2,6-bisphosphatase: isozymic form, expression, and ...
|
|
32717
|
Enzymic hydrolysis of phosphonate esters. Reaction mechanism of intestinal 5'-nucleotide phosphodiesterase
|
|
32718
|
Enzymic hydrolysis of phosphonate esters. Reaction mechanism of intestinal 5'-nucleotide phosphodiesterase
|
|
32719
|
Enzymic hydrolysis of phosphonate esters. Reaction mechanism of intestinal 5'-nucleotide phosphodiesterase
|
|
32720
|
Enzymic hydrolysis of phosphonate esters. Reaction mechanism of intestinal 5'-nucleotide phosphodiesterase
|
|
32721
|
Enzymic hydrolysis of phosphonate esters. Reaction mechanism of intestinal 5'-nucleotide phosphodiesterase
|
|
32722
|
Enzymic hydrolysis of phosphonate esters. Reaction mechanism of intestinal 5'-nucleotide phosphodiesterase
|
|
32723
|
Enzymic hydrolysis of phosphonate esters. Reaction mechanism of intestinal 5'-nucleotide phosphodiesterase
|
|
32724
|
Purification and characterization of glucose oxidase from ligninolytic cultures of Phanerochaete chrysosporium
|
|
32725
|
Purification and characterization of glucose oxidase from ligninolytic cultures of Phanerochaete chrysosporium
|
|
32726
|
Purification and characterization of glucose oxidase from ligninolytic cultures of Phanerochaete chrysosporium
|
|
32727
|
Purification and characterization of glucose oxidase from ligninolytic cultures of Phanerochaete chrysosporium
|
|
32728
|
Purification and characterization of glucose oxidase from ligninolytic cultures of Phanerochaete chrysosporium
|
|
32729
|
NADPH-cytochrome c reductase of Candida tropicalis grown on alkane
|
|
32730
|
NADPH-cytochrome c reductase of Candida tropicalis grown on alkane
|
|
32731
|
NADPH-cytochrome c reductase of Candida tropicalis grown on alkane
|
|
32732
|
NADPH-cytochrome c reductase of Candida tropicalis grown on alkane
|
|
32733
|
NADPH-cytochrome c reductase of Candida tropicalis grown on alkane
|
|
32734
|
NADPH-cytochrome c reductase of Candida tropicalis grown on alkane
|
|
32735
|
NADPH-cytochrome c reductase of Candida tropicalis grown on alkane
|
|
32736
|
Ornithine oxoacid aminotransferase found in AH 130 ascites hepatoma cells
|
|
32737
|
Ornithine oxoacid aminotransferase found in AH 130 ascites hepatoma cells
|
|
32738
|
Ornithine oxoacid aminotransferase found in AH 130 ascites hepatoma cells
|
|
32739
|
Ornithine oxoacid aminotransferase found in AH 130 ascites hepatoma cells
|
|
32740
|
Ornithine oxoacid aminotransferase found in AH 130 ascites hepatoma cells
|
|
32741
|
Ornithine oxoacid aminotransferase found in AH 130 ascites hepatoma cells
|
|
32742
|
Ornithine oxoacid aminotransferase found in AH 130 ascites hepatoma cells
|
|
32743
|
Ornithine oxoacid aminotransferase found in AH 130 ascites hepatoma cells
|
|
32744
|
Ornithine oxoacid aminotransferase found in AH 130 ascites hepatoma cells
|
|
32745
|
Ornithine oxoacid aminotransferase found in AH 130 ascites hepatoma cells
|
|
32746
|
Ornithine oxoacid aminotransferase found in AH 130 ascites hepatoma cells
|
|
32747
|
Ornithine oxoacid aminotransferase found in AH 130 ascites hepatoma cells
|
|
32748
|
Ornithine oxoacid aminotransferase found in AH 130 ascites hepatoma cells
|
|
32749
|
Ornithine oxoacid aminotransferase found in AH 130 ascites hepatoma cells
|
|
32750
|
Ornithine oxoacid aminotransferase found in AH 130 ascites hepatoma cells
|
|
32751
|
Ornithine oxoacid aminotransferase found in AH 130 ascites hepatoma cells
|
|
32752
|
Functional importance of Asp56 from the alpha-polypeptide of Phaseolus vulgaris glutamine synthetase. An ...
|
|
32753
|
Functional importance of Asp56 from the alpha-polypeptide of Phaseolus vulgaris glutamine synthetase. An ...
|
|
32754
|
Functional importance of Asp56 from the alpha-polypeptide of Phaseolus vulgaris glutamine synthetase. An ...
|
|
32755
|
Functional importance of Asp56 from the alpha-polypeptide of Phaseolus vulgaris glutamine synthetase. An ...
|
|
32756
|
Functional importance of Asp56 from the alpha-polypeptide of Phaseolus vulgaris glutamine synthetase. An ...
|
|
32757
|
Functional importance of Asp56 from the alpha-polypeptide of Phaseolus vulgaris glutamine synthetase. An ...
|
|
32758
|
Functional importance of Asp56 from the alpha-polypeptide of Phaseolus vulgaris glutamine synthetase. An ...
|
|
32759
|
Functional importance of Asp56 from the alpha-polypeptide of Phaseolus vulgaris glutamine synthetase. An ...
|
|
32760
|
Functional importance of Asp56 from the alpha-polypeptide of Phaseolus vulgaris glutamine synthetase. An ...
|
|
32761
|
Functional importance of Asp56 from the alpha-polypeptide of Phaseolus vulgaris glutamine synthetase. An ...
|
|
32762
|
Functional importance of Asp56 from the alpha-polypeptide of Phaseolus vulgaris glutamine synthetase. An ...
|
|
32763
|
Functional importance of Asp56 from the alpha-polypeptide of Phaseolus vulgaris glutamine synthetase. An ...
|
|
32764
|
Molecular characterization of a glucokinase with broad hexose specificity from Bacillus sphaericus strain ...
|
|
32765
|
Molecular characterization of a glucokinase with broad hexose specificity from Bacillus sphaericus strain ...
|
|
32766
|
Molecular characterization of a glucokinase with broad hexose specificity from Bacillus sphaericus strain ...
|
|
32767
|
Important role of Ser443 in different thermal stability of human glutamate dehydrogenase isozymes
|
|
32768
|
Important role of Ser443 in different thermal stability of human glutamate dehydrogenase isozymes
|
|
32769
|
Important role of Ser443 in different thermal stability of human glutamate dehydrogenase isozymes
|
|
32770
|
Important role of Ser443 in different thermal stability of human glutamate dehydrogenase isozymes
|
|
32771
|
Important role of Ser443 in different thermal stability of human glutamate dehydrogenase isozymes
|
|
32772
|
Important role of Ser443 in different thermal stability of human glutamate dehydrogenase isozymes
|
|
32773
|
citrate inhibition-resistant form of 6-phosphofructo-1-kinase from Aspergillus niger
|
|
32774
|
citrate inhibition-resistant form of 6-phosphofructo-1-kinase from Aspergillus niger
|
|
32775
|
citrate inhibition-resistant form of 6-phosphofructo-1-kinase from Aspergillus niger
|
|
32776
|
A small, thermostable, and monofunctional chorismate mutase from the archaeon Methanococcus jannaschii.
|
|
32777
|
A small, thermostable, and monofunctional chorismate mutase from the archaeon Methanococcus jannaschii.
|
|
32778
|
Purification, microsequencing and cloning of spinach ATP-dependent phosphofructokinase link sequence and ...
|
|
32779
|
Purification, microsequencing and cloning of spinach ATP-dependent phosphofructokinase link sequence and ...
|
|
32780
|
Inhibition and inactivation of glucose-phosphorylating enzymes from Saccharomyces cerevisiae by D-xylose
|
|
32781
|
Inhibition and inactivation of glucose-phosphorylating enzymes from Saccharomyces cerevisiae by D-xylose
|
|
32782
|
Inhibition and inactivation of glucose-phosphorylating enzymes from Saccharomyces cerevisiae by D-xylose
|
|
32783
|
Inhibition and inactivation of glucose-phosphorylating enzymes from Saccharomyces cerevisiae by D-xylose
|
|
32784
|
Inhibition and inactivation of glucose-phosphorylating enzymes from Saccharomyces cerevisiae by D-xylose
|
|
32785
|
Inhibition and inactivation of glucose-phosphorylating enzymes from Saccharomyces cerevisiae by D-xylose
|
|
32786
|
Inhibition and inactivation of glucose-phosphorylating enzymes from Saccharomyces cerevisiae by D-xylose
|
|
32787
|
Inhibition and inactivation of glucose-phosphorylating enzymes from Saccharomyces cerevisiae by D-xylose
|
|
32788
|
Inhibition and inactivation of glucose-phosphorylating enzymes from Saccharomyces cerevisiae by D-xylose
|
|
32789
|
Inhibition and inactivation of glucose-phosphorylating enzymes from Saccharomyces cerevisiae by D-xylose
|
|
32790
|
Identification and characterization of an ATP-dependent hexokinase with broad substrate specificity from the ...
|
|
32791
|
Identification and characterization of an ATP-dependent hexokinase with broad substrate specificity from the ...
|
|
32792
|
Identification and characterization of an ATP-dependent hexokinase with broad substrate specificity from the ...
|
|
32793
|
Identification and characterization of an ATP-dependent hexokinase with broad substrate specificity from the ...
|
|
32794
|
Identification and characterization of an ATP-dependent hexokinase with broad substrate specificity from the ...
|
|
32795
|
Identification and characterization of an ATP-dependent hexokinase with broad substrate specificity from the ...
|
|
32796
|
Identification and characterization of an ATP-dependent hexokinase with broad substrate specificity from the ...
|
|
32797
|
Identification and characterization of an ATP-dependent hexokinase with broad substrate specificity from the ...
|
|
32798
|
Identification and characterization of an ATP-dependent hexokinase with broad substrate specificity from the ...
|
|
32799
|
The Botrytis cinerea hexokinase, Hxk1, but not the glucokinase, Glk1, is required for normal growth and sugar ...
|
|
32800
|
The Botrytis cinerea hexokinase, Hxk1, but not the glucokinase, Glk1, is required for normal growth and sugar ...
|
|
32801
|
The Botrytis cinerea hexokinase, Hxk1, but not the glucokinase, Glk1, is required for normal growth and sugar ...
|
|
32802
|
Human aldehyde dehydrogenase: metabolism of putrescine and histamine
|
|
32803
|
Human aldehyde dehydrogenase: metabolism of putrescine and histamine
|
|
32804
|
Human aldehyde dehydrogenase: metabolism of putrescine and histamine
|
|
32805
|
Human aldehyde dehydrogenase: metabolism of putrescine and histamine
|
|
32806
|
Human aldehyde dehydrogenase: metabolism of putrescine and histamine
|
|
32807
|
Human aldehyde dehydrogenase: metabolism of putrescine and histamine
|
|
32808
|
Human aldehyde dehydrogenase: metabolism of putrescine and histamine
|
|
32809
|
Human aldehyde dehydrogenase: metabolism of putrescine and histamine
|
|
32810
|
Human aldehyde dehydrogenase: metabolism of putrescine and histamine
|
|
32811
|
Human aldehyde dehydrogenase: metabolism of putrescine and histamine
|
|
32812
|
Human aldehyde dehydrogenase: metabolism of putrescine and histamine
|
|
32813
|
Human aldehyde dehydrogenase: metabolism of putrescine and histamine
|
|
32814
|
Human aldehyde dehydrogenase: metabolism of putrescine and histamine
|
|
32815
|
Human aldehyde dehydrogenase: metabolism of putrescine and histamine
|
|
32816
|
Human aldehyde dehydrogenase: metabolism of putrescine and histamine
|
|
32817
|
Human aldehyde dehydrogenase: metabolism of putrescine and histamine
|
|
32818
|
Human aldehyde dehydrogenase: metabolism of putrescine and histamine
|
|
32819
|
Human aldehyde dehydrogenase: metabolism of putrescine and histamine
|
|
32820
|
Human aldehyde dehydrogenase: metabolism of putrescine and histamine
|
|
32821
|
Human aldehyde dehydrogenase: metabolism of putrescine and histamine
|
|
32822
|
Human aldehyde dehydrogenase: metabolism of putrescine and histamine
|
|
32823
|
New highly selective inhibitors of class II fructose-1,6-bisphosphate aldolases
|
|
32824
|
New highly selective inhibitors of class II fructose-1,6-bisphosphate aldolases
|
|
32825
|
New highly selective inhibitors of class II fructose-1,6-bisphosphate aldolases
|
|
32826
|
New highly selective inhibitors of class II fructose-1,6-bisphosphate aldolases
|
|
32827
|
New highly selective inhibitors of class II fructose-1,6-bisphosphate aldolases
|
|
32828
|
New highly selective inhibitors of class II fructose-1,6-bisphosphate aldolases
|
|
32829
|
New highly selective inhibitors of class II fructose-1,6-bisphosphate aldolases
|
|
32830
|
Aldehyde dehydrogenase. Covalent intermediate in aldehyde dehydrogenation and ester hydrolysis
|
|
32831
|
Aldehyde dehydrogenase. Covalent intermediate in aldehyde dehydrogenation and ester hydrolysis
|
|
32832
|
Aldehyde dehydrogenase. Covalent intermediate in aldehyde dehydrogenation and ester hydrolysis
|
|
32833
|
Aldehyde dehydrogenase. Covalent intermediate in aldehyde dehydrogenation and ester hydrolysis
|
|
32834
|
Aldehyde dehydrogenase. Covalent intermediate in aldehyde dehydrogenation and ester hydrolysis
|
|
32835
|
Aldehyde dehydrogenase. Covalent intermediate in aldehyde dehydrogenation and ester hydrolysis
|
|
32836
|
Aldehyde dehydrogenase. Covalent intermediate in aldehyde dehydrogenation and ester hydrolysis
|
|
32837
|
Aldehyde dehydrogenase. Covalent intermediate in aldehyde dehydrogenation and ester hydrolysis
|
|
32838
|
Aldehyde dehydrogenase. Covalent intermediate in aldehyde dehydrogenation and ester hydrolysis
|
|
32839
|
Aldehyde dehydrogenase. Covalent intermediate in aldehyde dehydrogenation and ester hydrolysis
|
|
32840
|
Substrate specificity of bovine liver formaldehyde dehydrogenase
|
|
32841
|
Substrate specificity of bovine liver formaldehyde dehydrogenase
|
|
32842
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32843
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32844
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32845
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32846
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32847
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32848
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32849
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32850
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32851
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32852
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32853
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32854
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32855
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32856
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32857
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32858
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32859
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32860
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32861
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32862
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32863
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32864
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32865
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32866
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32867
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32868
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32869
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32870
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32871
|
UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human ...
|
|
32872
|
Heterologous expression, purification, and kinetic comparison of the cytoplasmic and mitochondrial glyoxalase ...
|
|
32873
|
Heterologous expression, purification, and kinetic comparison of the cytoplasmic and mitochondrial glyoxalase ...
|
|
32874
|
Heterologous expression, purification, and kinetic comparison of the cytoplasmic and mitochondrial glyoxalase ...
|
|
32875
|
Heterologous expression, purification, and kinetic comparison of the cytoplasmic and mitochondrial glyoxalase ...
|
|
32876
|
Heterologous expression, purification, and kinetic comparison of the cytoplasmic and mitochondrial glyoxalase ...
|
|
32877
|
Heterologous expression, purification, and kinetic comparison of the cytoplasmic and mitochondrial glyoxalase ...
|
|
32878
|
Heterologous expression, purification, and kinetic comparison of the cytoplasmic and mitochondrial glyoxalase ...
|
|
32879
|
Heterologous expression, purification, and kinetic comparison of the cytoplasmic and mitochondrial glyoxalase ...
|
|
32880
|
The tRNA-interacting factor p43 associates with mammalian arginyl-tRNA synthetase but does not modify its tRNA ...
|
|
32881
|
The tRNA-interacting factor p43 associates with mammalian arginyl-tRNA synthetase but does not modify its tRNA ...
|
|
32882
|
The tRNA-interacting factor p43 associates with mammalian arginyl-tRNA synthetase but does not modify its tRNA ...
|
|
32883
|
The tRNA-interacting factor p43 associates with mammalian arginyl-tRNA synthetase but does not modify its tRNA ...
|
|
32884
|
The tRNA-interacting factor p43 associates with mammalian arginyl-tRNA synthetase but does not modify its tRNA ...
|
|
32885
|
The tRNA-interacting factor p43 associates with mammalian arginyl-tRNA synthetase but does not modify its tRNA ...
|
|
32886
|
The tRNA-interacting factor p43 associates with mammalian arginyl-tRNA synthetase but does not modify its tRNA ...
|
|
32887
|
The tRNA-interacting factor p43 associates with mammalian arginyl-tRNA synthetase but does not modify its tRNA ...
|
|
32888
|
Biochemical and mass spectrometric characterization of soluble ecto-5'-nucleotidase from bull seminal plasma
|
|
32889
|
Biochemical and mass spectrometric characterization of soluble ecto-5'-nucleotidase from bull seminal plasma
|
|
32890
|
Biochemical and mass spectrometric characterization of soluble ecto-5'-nucleotidase from bull seminal plasma
|
|
32891
|
Biochemical and mass spectrometric characterization of soluble ecto-5'-nucleotidase from bull seminal plasma
|
|
32892
|
Biochemical and mass spectrometric characterization of soluble ecto-5'-nucleotidase from bull seminal plasma
|
|
32893
|
Biochemical and mass spectrometric characterization of soluble ecto-5'-nucleotidase from bull seminal plasma
|
|
32894
|
Biochemical and mass spectrometric characterization of soluble ecto-5'-nucleotidase from bull seminal plasma
|
|
32895
|
Biochemical and mass spectrometric characterization of soluble ecto-5'-nucleotidase from bull seminal plasma
|
|
32896
|
Biochemical and mass spectrometric characterization of soluble ecto-5'-nucleotidase from bull seminal plasma
|
|
32897
|
Biochemical and mass spectrometric characterization of soluble ecto-5'-nucleotidase from bull seminal plasma
|
|
32898
|
Biochemical and mass spectrometric characterization of soluble ecto-5'-nucleotidase from bull seminal plasma
|
|
32899
|
Biochemical and mass spectrometric characterization of soluble ecto-5'-nucleotidase from bull seminal plasma
|
|
32900
|
Biochemical and mass spectrometric characterization of soluble ecto-5'-nucleotidase from bull seminal plasma
|
|
32901
|
Biochemical and mass spectrometric characterization of soluble ecto-5'-nucleotidase from bull seminal plasma
|
|
32902
|
Biochemical and mass spectrometric characterization of soluble ecto-5'-nucleotidase from bull seminal plasma
|
|
32903
|
Biochemical and mass spectrometric characterization of soluble ecto-5'-nucleotidase from bull seminal plasma
|
|
32904
|
The suicide inactivation of ox liver short-chain acyl-CoA dehydrogenase by propionyl-CoA. Formation of an FAD ...
|
|
32905
|
The suicide inactivation of ox liver short-chain acyl-CoA dehydrogenase by propionyl-CoA. Formation of an FAD ...
|
|
32906
|
The suicide inactivation of ox liver short-chain acyl-CoA dehydrogenase by propionyl-CoA. Formation of an FAD ...
|
|
32907
|
The suicide inactivation of ox liver short-chain acyl-CoA dehydrogenase by propionyl-CoA. Formation of an FAD ...
|
|
32908
|
The suicide inactivation of ox liver short-chain acyl-CoA dehydrogenase by propionyl-CoA. Formation of an FAD ...
|
|
32909
|
The suicide inactivation of ox liver short-chain acyl-CoA dehydrogenase by propionyl-CoA. Formation of an FAD ...
|
|
32910
|
The suicide inactivation of ox liver short-chain acyl-CoA dehydrogenase by propionyl-CoA. Formation of an FAD ...
|
|
32911
|
The suicide inactivation of ox liver short-chain acyl-CoA dehydrogenase by propionyl-CoA. Formation of an FAD ...
|
|
32912
|
The suicide inactivation of ox liver short-chain acyl-CoA dehydrogenase by propionyl-CoA. Formation of an FAD ...
|
|
32913
|
The suicide inactivation of ox liver short-chain acyl-CoA dehydrogenase by propionyl-CoA. Formation of an FAD ...
|
|
32914
|
Autocatalytic processing of pro-papaya proteinase IV is prevented by crowding of the active-site cleft
|
|
32915
|
Autocatalytic processing of pro-papaya proteinase IV is prevented by crowding of the active-site cleft
|
|
32916
|
Autocatalytic processing of pro-papaya proteinase IV is prevented by crowding of the active-site cleft
|
|
32917
|
Autocatalytic processing of pro-papaya proteinase IV is prevented by crowding of the active-site cleft
|
|
32918
|
Inhibition of the low-Km mitochondrial aldehyde dehydrogenase by diethyl maleate and phorone in vivo and in ...
|
|
32919
|
Inhibition of the low-Km mitochondrial aldehyde dehydrogenase by diethyl maleate and phorone in vivo and in ...
|
|
32920
|
Inhibition of the low-Km mitochondrial aldehyde dehydrogenase by diethyl maleate and phorone in vivo and in ...
|
|
32921
|
Inhibition of the low-Km mitochondrial aldehyde dehydrogenase by diethyl maleate and phorone in vivo and in ...
|
|
32922
|
Crystallization and properties of human liver ornithine aminotransferase
|
|
32923
|
Crystallization and properties of human liver ornithine aminotransferase
|
|
32924
|
Crystallization and properties of human liver ornithine aminotransferase
|
|
32925
|
Crystallization and properties of human liver ornithine aminotransferase
|
|
32926
|
Crystallization and properties of human liver ornithine aminotransferase
|
|
32927
|
Crystallization and properties of human liver ornithine aminotransferase
|
|
32928
|
Crystallization and properties of human liver ornithine aminotransferase
|
|
32929
|
Crystallization and properties of human liver ornithine aminotransferase
|
|
32930
|
Separation and properties of five distinct acyl-CoA dehydrogenases from rat liver mitochondria. Identification ...
|
|
32931
|
Separation and properties of five distinct acyl-CoA dehydrogenases from rat liver mitochondria. Identification ...
|
|
32932
|
Separation and properties of five distinct acyl-CoA dehydrogenases from rat liver mitochondria. Identification ...
|
|
32933
|
Separation and properties of five distinct acyl-CoA dehydrogenases from rat liver mitochondria. Identification ...
|
|
32934
|
Separation and properties of five distinct acyl-CoA dehydrogenases from rat liver mitochondria. Identification ...
|
|
32935
|
Separation and properties of five distinct acyl-CoA dehydrogenases from rat liver mitochondria. Identification ...
|
|
32936
|
Separation and properties of five distinct acyl-CoA dehydrogenases from rat liver mitochondria. Identification ...
|
|
32937
|
Separation and properties of five distinct acyl-CoA dehydrogenases from rat liver mitochondria. Identification ...
|
|
32938
|
Separation and properties of five distinct acyl-CoA dehydrogenases from rat liver mitochondria. Identification ...
|
|
32939
|
Separation and properties of five distinct acyl-CoA dehydrogenases from rat liver mitochondria. Identification ...
|
|
32940
|
Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from ...
|
|
32941
|
Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from ...
|
|
32942
|
Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from ...
|
|
32943
|
Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from ...
|
|
32944
|
Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from ...
|
|
32945
|
Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from ...
|
|
32946
|
Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from ...
|
|
32947
|
Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from ...
|
|
32948
|
Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from ...
|
|
32949
|
Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from ...
|
|
32950
|
Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from ...
|
|
32951
|
Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from ...
|
|
32952
|
Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from ...
|
|
32953
|
Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from ...
|
|
32954
|
Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from ...
|
|
32955
|
Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from ...
|
|
32956
|
Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from ...
|
|
32957
|
Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from ...
|
|
32958
|
Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from ...
|
|
32959
|
Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from ...
|
|
32960
|
Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from ...
|
|
32961
|
Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from ...
|
|
32962
|
Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from ...
|
|
32963
|
Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from ...
|
|
32964
|
Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from ...
|
|
32965
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32966
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32967
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32968
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32969
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32970
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32971
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32972
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32973
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32974
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32975
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32976
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32977
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32978
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32979
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32980
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32981
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32982
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32983
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32984
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32985
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32986
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32987
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32988
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32989
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32990
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32991
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32992
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32993
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32994
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32995
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32996
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32997
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32998
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
32999
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|
|
33000
|
Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases ...
|





 If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.
If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.