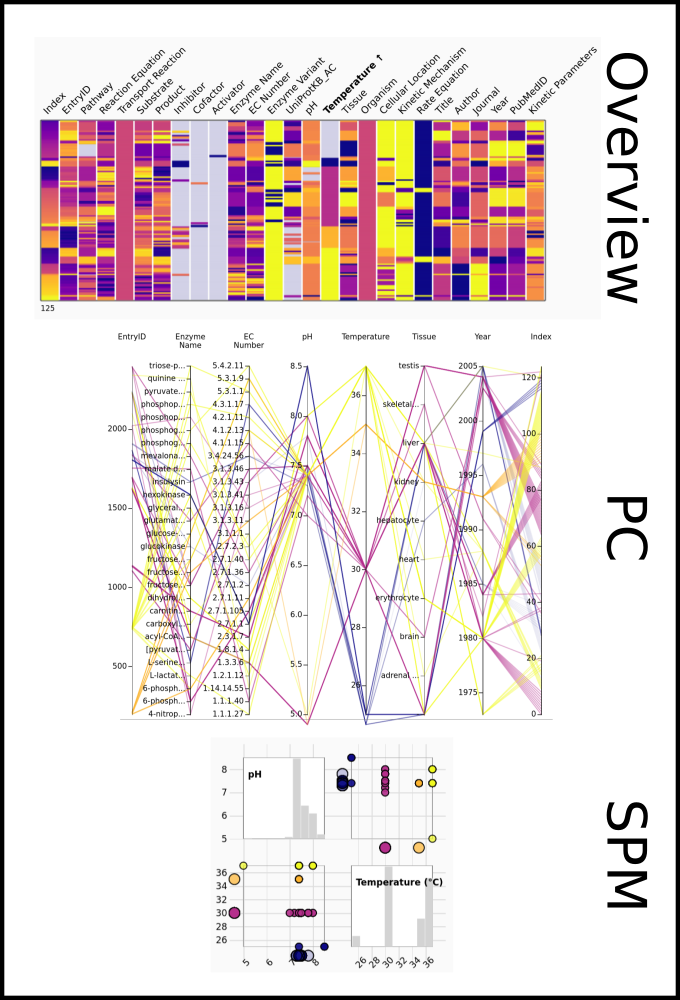
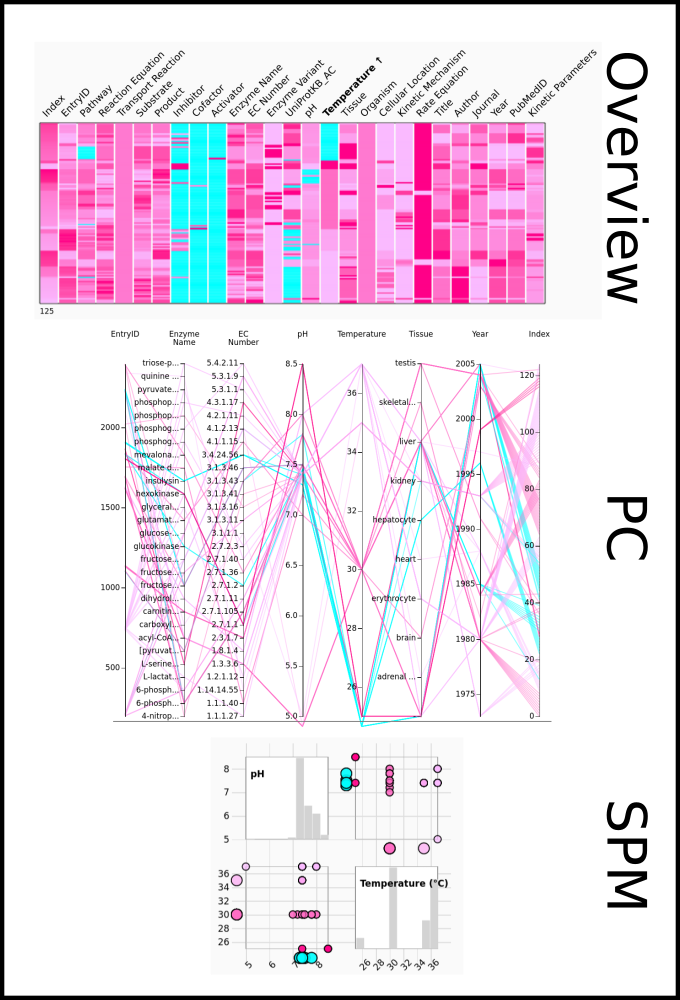
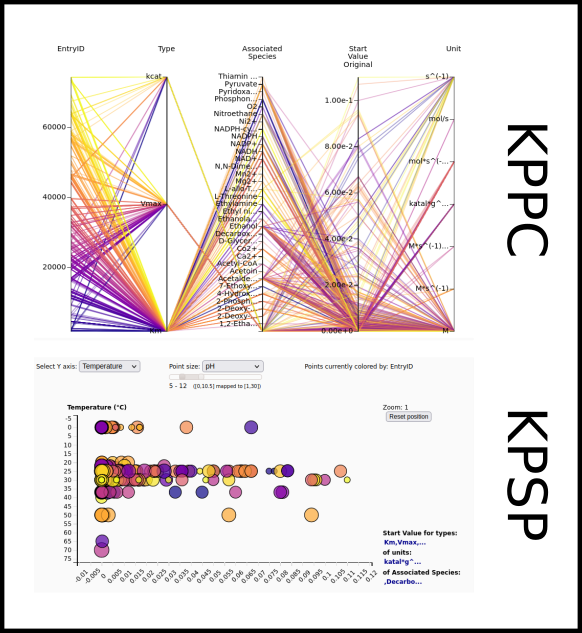
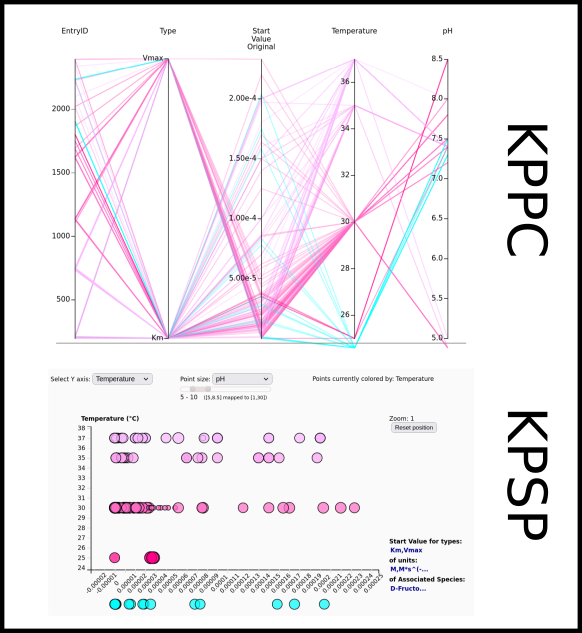
To improve SABIO-RK data interoperability, semantic markup was added to web pages as described and defined by
This structured information makes it easier to discover, collate and analyse our data.
|
3001
|
Cysteine and keto acids modulate mosquito kynurenine aminotransferase catalyzed kynurenic acid production
|
|
3002
|
Cysteine and keto acids modulate mosquito kynurenine aminotransferase catalyzed kynurenic acid production
|
|
3003
|
Cysteine and keto acids modulate mosquito kynurenine aminotransferase catalyzed kynurenic acid production
|
|
3004
|
Cysteine and keto acids modulate mosquito kynurenine aminotransferase catalyzed kynurenic acid production
|
|
3005
|
Cysteine and keto acids modulate mosquito kynurenine aminotransferase catalyzed kynurenic acid production
|
|
3006
|
Cysteine and keto acids modulate mosquito kynurenine aminotransferase catalyzed kynurenic acid production
|
|
3007
|
Cysteine and keto acids modulate mosquito kynurenine aminotransferase catalyzed kynurenic acid production
|
|
3008
|
Cysteine and keto acids modulate mosquito kynurenine aminotransferase catalyzed kynurenic acid production
|
|
3009
|
Cysteine and keto acids modulate mosquito kynurenine aminotransferase catalyzed kynurenic acid production
|
|
3010
|
Rat Liver gamma-Butyrobetaine Hydroxylase Catalyzed Reaction: Influence of Potassium, Substrates, and ...
|
|
3011
|
Rat Liver gamma-Butyrobetaine Hydroxylase Catalyzed Reaction: Influence of Potassium, Substrates, and ...
|
|
3012
|
Rat Liver gamma-Butyrobetaine Hydroxylase Catalyzed Reaction: Influence of Potassium, Substrates, and ...
|
|
3013
|
Rat Liver gamma-Butyrobetaine Hydroxylase Catalyzed Reaction: Influence of Potassium, Substrates, and ...
|
|
3014
|
Rat Liver gamma-Butyrobetaine Hydroxylase Catalyzed Reaction: Influence of Potassium, Substrates, and ...
|
|
3015
|
Rat Liver gamma-Butyrobetaine Hydroxylase Catalyzed Reaction: Influence of Potassium, Substrates, and ...
|
|
3016
|
Rat Liver gamma-Butyrobetaine Hydroxylase Catalyzed Reaction: Influence of Potassium, Substrates, and ...
|
|
3017
|
Rat Liver gamma-Butyrobetaine Hydroxylase Catalyzed Reaction: Influence of Potassium, Substrates, and ...
|
|
3018
|
Kinetics of the activation of rat liver pyruvate kinase by fructose 1,6-diphosphate and methods for ...
|
|
3019
|
Kinetics of the activation of rat liver pyruvate kinase by fructose 1,6-diphosphate and methods for ...
|
|
3020
|
Kinetics of the activation of rat liver pyruvate kinase by fructose 1,6-diphosphate and methods for ...
|
|
3021
|
Kinetics of the activation of rat liver pyruvate kinase by fructose 1,6-diphosphate and methods for ...
|
|
3022
|
Kinetics of the activation of rat liver pyruvate kinase by fructose 1,6-diphosphate and methods for ...
|
|
3023
|
Kinetics of the activation of rat liver pyruvate kinase by fructose 1,6-diphosphate and methods for ...
|
|
3024
|
Separation, Properties, and regulation of Acyl Coenzyme A dehydrogenase from Bovine Heart and Liver
|
|
3025
|
Separation, Properties, and regulation of Acyl Coenzyme A dehydrogenase from Bovine Heart and Liver
|
|
3026
|
Separation, Properties, and regulation of Acyl Coenzyme A dehydrogenase from Bovine Heart and Liver
|
|
3027
|
Separation, Properties, and regulation of Acyl Coenzyme A dehydrogenase from Bovine Heart and Liver
|
|
3028
|
Separation, Properties, and regulation of Acyl Coenzyme A dehydrogenase from Bovine Heart and Liver
|
|
3029
|
Separation, Properties, and regulation of Acyl Coenzyme A dehydrogenase from Bovine Heart and Liver
|
|
3030
|
Separation, Properties, and regulation of Acyl Coenzyme A dehydrogenase from Bovine Heart and Liver
|
|
3031
|
Separation, Properties, and regulation of Acyl Coenzyme A dehydrogenase from Bovine Heart and Liver
|
|
3032
|
Separation, Properties, and regulation of Acyl Coenzyme A dehydrogenase from Bovine Heart and Liver
|
|
3033
|
Separation, Properties, and regulation of Acyl Coenzyme A dehydrogenase from Bovine Heart and Liver
|
|
3034
|
Separation, Properties, and regulation of Acyl Coenzyme A dehydrogenase from Bovine Heart and Liver
|
|
3035
|
Separation, Properties, and regulation of Acyl Coenzyme A dehydrogenase from Bovine Heart and Liver
|
|
3036
|
Tyrosinase inhibitory activity of cucumber compounds: enzymes responsible for browning in cucumber
|
|
3037
|
Tyrosinase inhibitory activity of cucumber compounds: enzymes responsible for browning in cucumber
|
|
3038
|
Tyrosinase inhibitory activity of cucumber compounds: enzymes responsible for browning in cucumber
|
|
3039
|
Alpha-glucan, water dikinase (GWD): a plastidic enzyme with redox-regulated and coordinated catalytic activity ...
|
|
3040
|
Alpha-glucan, water dikinase (GWD): a plastidic enzyme with redox-regulated and coordinated catalytic activity ...
|
|
3041
|
Alpha-glucan, water dikinase (GWD): a plastidic enzyme with redox-regulated and coordinated catalytic activity ...
|
|
3042
|
Acetylcholinesterase active centre and gorge conformations analysed by combinatorial mutations and ...
|
|
3043
|
Acetylcholinesterase active centre and gorge conformations analysed by combinatorial mutations and ...
|
|
3044
|
Acetylcholinesterase active centre and gorge conformations analysed by combinatorial mutations and ...
|
|
3045
|
Acetylcholinesterase active centre and gorge conformations analysed by combinatorial mutations and ...
|
|
3046
|
Acetylcholinesterase active centre and gorge conformations analysed by combinatorial mutations and ...
|
|
3047
|
Acetylcholinesterase active centre and gorge conformations analysed by combinatorial mutations and ...
|
|
3048
|
Acetylcholinesterase active centre and gorge conformations analysed by combinatorial mutations and ...
|
|
3049
|
Acetylcholinesterase active centre and gorge conformations analysed by combinatorial mutations and ...
|
|
3050
|
Acetylcholinesterase active centre and gorge conformations analysed by combinatorial mutations and ...
|
|
3051
|
Acetylcholinesterase active centre and gorge conformations analysed by combinatorial mutations and ...
|
|
3052
|
Acetylcholinesterase active centre and gorge conformations analysed by combinatorial mutations and ...
|
|
3053
|
Acetylcholinesterase active centre and gorge conformations analysed by combinatorial mutations and ...
|
|
3054
|
Acetylcholinesterase active centre and gorge conformations analysed by combinatorial mutations and ...
|
|
3055
|
Acetylcholinesterase active centre and gorge conformations analysed by combinatorial mutations and ...
|
|
3056
|
Chemical mechanism and substrate specificity of RhlI, an acylhomoserine lactone synthase from Pseudomonas ...
|
|
3057
|
Chemical mechanism and substrate specificity of RhlI, an acylhomoserine lactone synthase from Pseudomonas ...
|
|
3058
|
Chemical mechanism and substrate specificity of RhlI, an acylhomoserine lactone synthase from Pseudomonas ...
|
|
3059
|
Chemical mechanism and substrate specificity of RhlI, an acylhomoserine lactone synthase from Pseudomonas ...
|
|
3060
|
MtmMII-mediated C-methylation during biosynthesis of the antitumor drug mithramycin is essential for ...
|
|
3061
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3062
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3063
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3064
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3065
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3066
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3067
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3068
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3069
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3070
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3071
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3072
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3073
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3074
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3075
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3076
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3077
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3078
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3079
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3080
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3081
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3082
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3083
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3084
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3085
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3086
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3087
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3088
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3089
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3090
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3091
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3092
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3093
|
Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7
|
|
3094
|
Mechanism of chorismate synthase. Role of the two invariant histidine residues in the active site
|
|
3095
|
Mechanism of chorismate synthase. Role of the two invariant histidine residues in the active site
|
|
3096
|
Mechanism of chorismate synthase. Role of the two invariant histidine residues in the active site
|
|
3097
|
Mechanism of chorismate synthase. Role of the two invariant histidine residues in the active site
|
|
3098
|
Mechanism of chorismate synthase. Role of the two invariant histidine residues in the active site
|
|
3099
|
Mechanism of chorismate synthase. Role of the two invariant histidine residues in the active site
|
|
3100
|
Mechanism of chorismate synthase. Role of the two invariant histidine residues in the active site
|
|
3101
|
Redox regulation and reaction mechanism of human cystathionine-beta-synthase: a PLP-dependent hemesensor ...
|
|
3102
|
Redox regulation and reaction mechanism of human cystathionine-beta-synthase: a PLP-dependent hemesensor ...
|
|
3103
|
Redox regulation and reaction mechanism of human cystathionine-beta-synthase: a PLP-dependent hemesensor ...
|
|
3104
|
Probing hydrogen-bonding interactions in the active site of medium-chain acyl-CoA dehydrogenase using Raman ...
|
|
3105
|
Probing hydrogen-bonding interactions in the active site of medium-chain acyl-CoA dehydrogenase using Raman ...
|
|
3106
|
Probing hydrogen-bonding interactions in the active site of medium-chain acyl-CoA dehydrogenase using Raman ...
|
|
3107
|
Probing hydrogen-bonding interactions in the active site of medium-chain acyl-CoA dehydrogenase using Raman ...
|
|
3108
|
Probing hydrogen-bonding interactions in the active site of medium-chain acyl-CoA dehydrogenase using Raman ...
|
|
3141
|
Studies on Bovine Hepatic Fructose 1,6-Diphosphatase
|
|
3142
|
Studies on Bovine Hepatic Fructose 1,6-Diphosphatase
|
|
3143
|
Characterization of the peroxidase system at low H2O2
|
|
3144
|
Characterization of the peroxidase system at low H2O2
|
|
3145
|
Characterization of the peroxidase system at low H2O2
|
|
3146
|
Characterization of the peroxidase system at low H2O2
|
|
3147
|
Characterization of the peroxidase system at low H2O2
|
|
3148
|
Modifying the stereochemistry of an enzyme-catalyzed reaction by directed evolution
|
|
3149
|
Modifying the stereochemistry of an enzyme-catalyzed reaction by directed evolution
|
|
3150
|
Modifying the stereochemistry of an enzyme-catalyzed reaction by directed evolution
|
|
3151
|
Modifying the stereochemistry of an enzyme-catalyzed reaction by directed evolution
|
|
3152
|
Modifying the stereochemistry of an enzyme-catalyzed reaction by directed evolution
|
|
3153
|
Modifying the stereochemistry of an enzyme-catalyzed reaction by directed evolution
|
|
3154
|
Modifying the stereochemistry of an enzyme-catalyzed reaction by directed evolution
|
|
3155
|
Modifying the stereochemistry of an enzyme-catalyzed reaction by directed evolution
|
|
3156
|
Modifying the stereochemistry of an enzyme-catalyzed reaction by directed evolution
|
|
3157
|
Modifying the stereochemistry of an enzyme-catalyzed reaction by directed evolution
|
|
3158
|
Modifying the stereochemistry of an enzyme-catalyzed reaction by directed evolution
|
|
3159
|
Modifying the stereochemistry of an enzyme-catalyzed reaction by directed evolution
|
|
3160
|
Modifying the stereochemistry of an enzyme-catalyzed reaction by directed evolution
|
|
3161
|
Modifying the stereochemistry of an enzyme-catalyzed reaction by directed evolution
|
|
3187
|
Inhibition of lentil copper/TPQ amine oxidase by the mechanism-based inhibitor derived from tyramine
|
|
3188
|
Inhibition of lentil copper/TPQ amine oxidase by the mechanism-based inhibitor derived from tyramine
|
|
3189
|
Inhibition of lentil copper/TPQ amine oxidase by the mechanism-based inhibitor derived from tyramine
|
|
3190
|
Inhibition of lentil copper/TPQ amine oxidase by the mechanism-based inhibitor derived from tyramine
|
|
3191
|
Inhibition of lentil copper/TPQ amine oxidase by the mechanism-based inhibitor derived from tyramine
|
|
3192
|
Inhibition of lentil copper/TPQ amine oxidase by the mechanism-based inhibitor derived from tyramine
|
|
3193
|
Kinetic studies of oxygen reactivity in soybean lipoxygenase-1
|
|
3194
|
Kinetic studies of oxygen reactivity in soybean lipoxygenase-1
|
|
3195
|
Kinetic studies of oxygen reactivity in soybean lipoxygenase-1
|
|
3196
|
Kinetic studies of oxygen reactivity in soybean lipoxygenase-1
|
|
3197
|
Kinetic studies of oxygen reactivity in soybean lipoxygenase-1
|
|
3198
|
Catalytic properties of mutant 23 S ribosomes resistant to oxazolidinones
|
|
3199
|
Catalytic properties of mutant 23 S ribosomes resistant to oxazolidinones
|
|
3200
|
Catalytic properties of mutant 23 S ribosomes resistant to oxazolidinones
|
|
3201
|
Catalytic properties of mutant 23 S ribosomes resistant to oxazolidinones
|
|
3202
|
Catalytic properties of mutant 23 S ribosomes resistant to oxazolidinones
|
|
3203
|
Catalytic properties of mutant 23 S ribosomes resistant to oxazolidinones
|
|
3204
|
Catalytic properties of mutant 23 S ribosomes resistant to oxazolidinones
|
|
3205
|
Catalytic properties of mutant 23 S ribosomes resistant to oxazolidinones
|
|
3206
|
Catalytic properties of mutant 23 S ribosomes resistant to oxazolidinones
|
|
3207
|
Catalytic properties of mutant 23 S ribosomes resistant to oxazolidinones
|
|
3208
|
Catalytic properties of mutant 23 S ribosomes resistant to oxazolidinones
|
|
3209
|
Catalytic properties of mutant 23 S ribosomes resistant to oxazolidinones
|
|
3210
|
Catalytic properties of mutant 23 S ribosomes resistant to oxazolidinones
|
|
3211
|
Catalytic properties of mutant 23 S ribosomes resistant to oxazolidinones
|
|
3212
|
Catalytic properties of mutant 23 S ribosomes resistant to oxazolidinones
|
|
3213
|
Catalytic properties of mutant 23 S ribosomes resistant to oxazolidinones
|
|
3214
|
Catalytic properties of mutant 23 S ribosomes resistant to oxazolidinones
|
|
3215
|
Catalytic properties of mutant 23 S ribosomes resistant to oxazolidinones
|
|
3216
|
Catalytic properties of mutant 23 S ribosomes resistant to oxazolidinones
|
|
3217
|
Allosteric and catalytic functions of the PPi-binding motif in the ATP sulfurylase-GTPase system
|
|
3218
|
Allosteric and catalytic functions of the PPi-binding motif in the ATP sulfurylase-GTPase system
|
|
3219
|
Allosteric and catalytic functions of the PPi-binding motif in the ATP sulfurylase-GTPase system
|
|
3220
|
Allosteric and catalytic functions of the PPi-binding motif in the ATP sulfurylase-GTPase system
|
|
3221
|
Allosteric and catalytic functions of the PPi-binding motif in the ATP sulfurylase-GTPase system
|
|
3222
|
Allosteric and catalytic functions of the PPi-binding motif in the ATP sulfurylase-GTPase system
|
|
3223
|
Allosteric and catalytic functions of the PPi-binding motif in the ATP sulfurylase-GTPase system
|
|
3224
|
Allosteric and catalytic functions of the PPi-binding motif in the ATP sulfurylase-GTPase system
|
|
3225
|
Allosteric and catalytic functions of the PPi-binding motif in the ATP sulfurylase-GTPase system
|
|
3226
|
Allosteric and catalytic functions of the PPi-binding motif in the ATP sulfurylase-GTPase system
|
|
3227
|
Allosteric and catalytic functions of the PPi-binding motif in the ATP sulfurylase-GTPase system
|
|
3228
|
Allosteric and catalytic functions of the PPi-binding motif in the ATP sulfurylase-GTPase system
|
|
3229
|
Allosteric and catalytic functions of the PPi-binding motif in the ATP sulfurylase-GTPase system
|
|
3230
|
Allosteric and catalytic functions of the PPi-binding motif in the ATP sulfurylase-GTPase system
|
|
3231
|
Relaxing the nicotinamide cofactor specificity of phosphite dehydrogenase by rational design
|
|
3232
|
Relaxing the nicotinamide cofactor specificity of phosphite dehydrogenase by rational design
|
|
3233
|
Relaxing the nicotinamide cofactor specificity of phosphite dehydrogenase by rational design
|
|
3234
|
Relaxing the nicotinamide cofactor specificity of phosphite dehydrogenase by rational design
|
|
3235
|
Relaxing the nicotinamide cofactor specificity of phosphite dehydrogenase by rational design
|
|
3236
|
Relaxing the nicotinamide cofactor specificity of phosphite dehydrogenase by rational design
|
|
3237
|
Relaxing the nicotinamide cofactor specificity of phosphite dehydrogenase by rational design
|
|
3238
|
Relaxing the nicotinamide cofactor specificity of phosphite dehydrogenase by rational design
|
|
3239
|
Characterization and kinetics of 45 kDa chitosanase from Bacillus sp. P16
|
|
3240
|
Escherichia coli cystathionine gamma-synthase does not obey ping-pong kinetics. Novel continuous assays for ...
|
|
3241
|
Escherichia coli cystathionine gamma-synthase does not obey ping-pong kinetics. Novel continuous assays for ...
|
|
3242
|
Escherichia coli cystathionine gamma-synthase does not obey ping-pong kinetics. Novel continuous assays for ...
|
|
3243
|
Escherichia coli cystathionine gamma-synthase does not obey ping-pong kinetics. Novel continuous assays for ...
|
|
3244
|
Escherichia coli cystathionine gamma-synthase does not obey ping-pong kinetics. Novel continuous assays for ...
|
|
3245
|
Escherichia coli cystathionine gamma-synthase does not obey ping-pong kinetics. Novel continuous assays for ...
|
|
3246
|
Escherichia coli cystathionine gamma-synthase does not obey ping-pong kinetics. Novel continuous assays for ...
|
|
3247
|
Escherichia coli cystathionine gamma-synthase does not obey ping-pong kinetics. Novel continuous assays for ...
|
|
3248
|
Escherichia coli cystathionine gamma-synthase does not obey ping-pong kinetics. Novel continuous assays for ...
|
|
3249
|
Escherichia coli cystathionine gamma-synthase does not obey ping-pong kinetics. Novel continuous assays for ...
|
|
3250
|
Potassium transport in a halophilic member of the bacteria domain: identification and characterization of the ...
|
|
3251
|
Potassium transport in a halophilic member of the bacteria domain: identification and characterization of the ...
|
|
3252
|
Potassium transport in a halophilic member of the bacteria domain: identification and characterization of the ...
|
|
3253
|
Treponema denticola cystalysin catalyzes beta-desulfination of L-cysteine sulfinic acid and ...
|
|
3254
|
Treponema denticola cystalysin catalyzes beta-desulfination of L-cysteine sulfinic acid and ...
|
|
3255
|
Treponema denticola cystalysin catalyzes beta-desulfination of L-cysteine sulfinic acid and ...
|
|
3256
|
Analysis of the role of the active site residue Arg98 in the flavoprotein tryptophan 2-monooxygenase, a member ...
|
|
3257
|
Analysis of the role of the active site residue Arg98 in the flavoprotein tryptophan 2-monooxygenase, a member ...
|
|
3258
|
Analysis of the role of the active site residue Arg98 in the flavoprotein tryptophan 2-monooxygenase, a member ...
|
|
3259
|
Analysis of the role of the active site residue Arg98 in the flavoprotein tryptophan 2-monooxygenase, a member ...
|
|
3260
|
Analysis of the role of the active site residue Arg98 in the flavoprotein tryptophan 2-monooxygenase, a member ...
|
|
3261
|
Analysis of the role of the active site residue Arg98 in the flavoprotein tryptophan 2-monooxygenase, a member ...
|
|
3262
|
Analysis of the role of the active site residue Arg98 in the flavoprotein tryptophan 2-monooxygenase, a member ...
|
|
3263
|
Analysis of the role of the active site residue Arg98 in the flavoprotein tryptophan 2-monooxygenase, a member ...
|
|
3264
|
Analysis of the role of the active site residue Arg98 in the flavoprotein tryptophan 2-monooxygenase, a member ...
|
|
3265
|
Analysis of the role of the active site residue Arg98 in the flavoprotein tryptophan 2-monooxygenase, a member ...
|
|
3266
|
Analysis of the role of the active site residue Arg98 in the flavoprotein tryptophan 2-monooxygenase, a member ...
|
|
3267
|
Analysis of the role of the active site residue Arg98 in the flavoprotein tryptophan 2-monooxygenase, a member ...
|
|
3283
|
Fluorescence study of ligand binding to potato tuber pyrophosphate-dependent phosphofructokinase: evidence for ...
|
|
3284
|
Fluorescence study of ligand binding to potato tuber pyrophosphate-dependent phosphofructokinase: evidence for ...
|
|
3285
|
Fluorescence study of ligand binding to potato tuber pyrophosphate-dependent phosphofructokinase: evidence for ...
|
|
3286
|
Fluorescence study of ligand binding to potato tuber pyrophosphate-dependent phosphofructokinase: evidence for ...
|
|
3287
|
Fluorescence study of ligand binding to potato tuber pyrophosphate-dependent phosphofructokinase: evidence for ...
|
|
3288
|
Fluorescence study of ligand binding to potato tuber pyrophosphate-dependent phosphofructokinase: evidence for ...
|
|
3289
|
Fluorescence study of ligand binding to potato tuber pyrophosphate-dependent phosphofructokinase: evidence for ...
|
|
3290
|
Fluorescence study of ligand binding to potato tuber pyrophosphate-dependent phosphofructokinase: evidence for ...
|
|
3291
|
Fluorescence study of ligand binding to potato tuber pyrophosphate-dependent phosphofructokinase: evidence for ...
|
|
3292
|
Fluorescence study of ligand binding to potato tuber pyrophosphate-dependent phosphofructokinase: evidence for ...
|
|
3293
|
Fluorescence study of ligand binding to potato tuber pyrophosphate-dependent phosphofructokinase: evidence for ...
|
|
3294
|
Fluorescence study of ligand binding to potato tuber pyrophosphate-dependent phosphofructokinase: evidence for ...
|
|
3295
|
Fluorescence study of ligand binding to potato tuber pyrophosphate-dependent phosphofructokinase: evidence for ...
|
|
3296
|
Fluorescence study of ligand binding to potato tuber pyrophosphate-dependent phosphofructokinase: evidence for ...
|
|
3297
|
Fluorescence study of ligand binding to potato tuber pyrophosphate-dependent phosphofructokinase: evidence for ...
|
|
3298
|
Fluorescence study of ligand binding to potato tuber pyrophosphate-dependent phosphofructokinase: evidence for ...
|
|
3299
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3300
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3301
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3302
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3303
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3304
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3305
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3306
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3307
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3308
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3309
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3310
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3311
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3312
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3313
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3314
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3315
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3316
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3317
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3318
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3319
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3320
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3321
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3322
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3323
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3324
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3325
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3326
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3327
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3328
|
Roles of low specificity and cofactor interaction sites on thrombin during factor XIII activation. Competition ...
|
|
3329
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3330
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3331
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3332
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3333
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3334
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3335
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3336
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3337
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3338
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3339
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3340
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3341
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3342
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3343
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3344
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3345
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3346
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3347
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3348
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3349
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3350
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3351
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3352
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3353
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3354
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3355
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3356
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3357
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3358
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3359
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3360
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3361
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3362
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3363
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3364
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3365
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3366
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3367
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3368
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3369
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3370
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3371
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3372
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3373
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3374
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3375
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3376
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3377
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3378
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3379
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3380
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3381
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3382
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3383
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3384
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3385
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3386
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3387
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3388
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3389
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3390
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3391
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3392
|
Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation ...
|
|
3393
|
Purification and properties of a new S-adenosyl-L-methionine:flavonoid 4`-O-methyltransferase from carnation ...
|
|
3394
|
Purification and properties of a new S-adenosyl-L-methionine:flavonoid 4`-O-methyltransferase from carnation ...
|
|
3395
|
Purification and properties of a new S-adenosyl-L-methionine:flavonoid 4`-O-methyltransferase from carnation ...
|
|
3396
|
Purification and properties of a new S-adenosyl-L-methionine:flavonoid 4`-O-methyltransferase from carnation ...
|
|
3397
|
The reverse activity of human acid ceramidase
|
|
3398
|
The reverse activity of human acid ceramidase
|
|
3399
|
Cuprous oxidase activity of yeast Fet3p and human ceruloplasmin: implication for function
|
|
3400
|
Cuprous oxidase activity of yeast Fet3p and human ceruloplasmin: implication for function
|
|
3401
|
Cuprous oxidase activity of yeast Fet3p and human ceruloplasmin: implication for function
|
|
3402
|
Cuprous oxidase activity of yeast Fet3p and human ceruloplasmin: implication for function
|
|
3403
|
Structural basis for catalytic differences between alpha class human glutathione transferases hGSTA1-1 and ...
|
|
3404
|
Structural basis for catalytic differences between alpha class human glutathione transferases hGSTA1-1 and ...
|
|
3405
|
Structural basis for catalytic differences between alpha class human glutathione transferases hGSTA1-1 and ...
|
|
3406
|
Structural basis for catalytic differences between alpha class human glutathione transferases hGSTA1-1 and ...
|
|
3407
|
Structural basis for catalytic differences between alpha class human glutathione transferases hGSTA1-1 and ...
|
|
3408
|
Structural basis for catalytic differences between alpha class human glutathione transferases hGSTA1-1 and ...
|
|
3409
|
Structural basis for catalytic differences between alpha class human glutathione transferases hGSTA1-1 and ...
|
|
3410
|
Structural basis for catalytic differences between alpha class human glutathione transferases hGSTA1-1 and ...
|
|
3411
|
Structural basis for catalytic differences between alpha class human glutathione transferases hGSTA1-1 and ...
|
|
3412
|
Structural basis for catalytic differences between alpha class human glutathione transferases hGSTA1-1 and ...
|
|
3413
|
Structural basis for catalytic differences between alpha class human glutathione transferases hGSTA1-1 and ...
|
|
3414
|
Structural basis for catalytic differences between alpha class human glutathione transferases hGSTA1-1 and ...
|
|
3415
|
Structural basis for catalytic differences between alpha class human glutathione transferases hGSTA1-1 and ...
|
|
3416
|
Structural basis for catalytic differences between alpha class human glutathione transferases hGSTA1-1 and ...
|
|
3417
|
Structural basis for catalytic differences between alpha class human glutathione transferases hGSTA1-1 and ...
|
|
3418
|
Structural basis for catalytic differences between alpha class human glutathione transferases hGSTA1-1 and ...
|
|
3419
|
Structural basis for catalytic differences between alpha class human glutathione transferases hGSTA1-1 and ...
|
|
3420
|
Structural basis for catalytic differences between alpha class human glutathione transferases hGSTA1-1 and ...
|
|
3421
|
Structural basis for catalytic differences between alpha class human glutathione transferases hGSTA1-1 and ...
|
|
3422
|
Structural basis for catalytic differences between alpha class human glutathione transferases hGSTA1-1 and ...
|
|
3423
|
Structural basis for catalytic differences between alpha class human glutathione transferases hGSTA1-1 and ...
|
|
3424
|
The cytosolic and glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei. Kinetic ...
|
|
3425
|
The cytosolic and glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei. Kinetic ...
|
|
3426
|
The cytosolic and glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei. Kinetic ...
|
|
3427
|
The cytosolic and glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei. Kinetic ...
|
|
3428
|
The cytosolic and glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei. Kinetic ...
|
|
3429
|
The cytosolic and glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei. Kinetic ...
|
|
3430
|
The cytosolic and glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei. Kinetic ...
|
|
3431
|
The cytosolic and glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei. Kinetic ...
|
|
3432
|
The cytosolic and glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei. Kinetic ...
|
|
3433
|
The cytosolic and glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei. Kinetic ...
|
|
3434
|
The cytosolic and glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei. Kinetic ...
|
|
3435
|
The cytosolic and glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei. Kinetic ...
|
|
3436
|
The cytosolic and glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei. Kinetic ...
|
|
3437
|
The cytosolic and glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei. Kinetic ...
|
|
3438
|
The cytosolic and glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei. Kinetic ...
|
|
3439
|
The cytosolic and glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei. Kinetic ...
|
|
3440
|
Human cytosolic 3alpha-hydroxysteroid dehydrogenases of the aldo-keto reductase superfamily display ...
|
|
3441
|
Human cytosolic 3alpha-hydroxysteroid dehydrogenases of the aldo-keto reductase superfamily display ...
|
|
3442
|
Human cytosolic 3alpha-hydroxysteroid dehydrogenases of the aldo-keto reductase superfamily display ...
|
|
3443
|
Human cytosolic 3alpha-hydroxysteroid dehydrogenases of the aldo-keto reductase superfamily display ...
|
|
3444
|
Human cytosolic 3alpha-hydroxysteroid dehydrogenases of the aldo-keto reductase superfamily display ...
|
|
3445
|
Human cytosolic 3alpha-hydroxysteroid dehydrogenases of the aldo-keto reductase superfamily display ...
|
|
3446
|
Human cytosolic 3alpha-hydroxysteroid dehydrogenases of the aldo-keto reductase superfamily display ...
|
|
3447
|
Human cytosolic 3alpha-hydroxysteroid dehydrogenases of the aldo-keto reductase superfamily display ...
|
|
3448
|
Human cytosolic 3alpha-hydroxysteroid dehydrogenases of the aldo-keto reductase superfamily display ...
|
|
3449
|
Human cytosolic 3alpha-hydroxysteroid dehydrogenases of the aldo-keto reductase superfamily display ...
|
|
3450
|
Human cytosolic 3alpha-hydroxysteroid dehydrogenases of the aldo-keto reductase superfamily display ...
|
|
3451
|
Human cytosolic 3alpha-hydroxysteroid dehydrogenases of the aldo-keto reductase superfamily display ...
|
|
3452
|
Human cytosolic 3alpha-hydroxysteroid dehydrogenases of the aldo-keto reductase superfamily display ...
|
|
3453
|
Human cytosolic 3alpha-hydroxysteroid dehydrogenases of the aldo-keto reductase superfamily display ...
|
|
3454
|
Human cytosolic 3alpha-hydroxysteroid dehydrogenases of the aldo-keto reductase superfamily display ...
|
|
3455
|
Human cytosolic 3alpha-hydroxysteroid dehydrogenases of the aldo-keto reductase superfamily display ...
|
|
3456
|
Control of glucose utilization in working perfused rat heart
|
|
3457
|
Control of glucose utilization in working perfused rat heart
|
|
3458
|
Control of glucose utilization in working perfused rat heart
|
|
3459
|
Control of glucose utilization in working perfused rat heart
|
|
3460
|
Control of glucose utilization in working perfused rat heart
|
|
3461
|
Control of glucose utilization in working perfused rat heart
|
|
3462
|
Control of glucose utilization in working perfused rat heart
|
|
3463
|
Control of glucose utilization in working perfused rat heart
|
|
3464
|
Control of glucose utilization in working perfused rat heart
|
|
3465
|
Control of glucose utilization in working perfused rat heart
|
|
3466
|
Control of glucose utilization in working perfused rat heart
|
|
3467
|
Control of glucose utilization in working perfused rat heart
|
|
3468
|
Control of glucose utilization in working perfused rat heart
|
|
3469
|
Control of glucose utilization in working perfused rat heart
|
|
3470
|
Control of glucose utilization in working perfused rat heart
|
|
3471
|
Control of glucose utilization in working perfused rat heart
|
|
3472
|
Control of glucose utilization in working perfused rat heart
|
|
3473
|
Control of glucose utilization in working perfused rat heart
|
|
3474
|
Control of glucose utilization in working perfused rat heart
|
|
3475
|
Control of glucose utilization in working perfused rat heart
|
|
3476
|
Control of glucose utilization in working perfused rat heart
|
|
3477
|
Control of glucose utilization in working perfused rat heart
|
|
3478
|
Control of glucose utilization in working perfused rat heart
|
|
3479
|
Control of glucose utilization in working perfused rat heart
|
|
3480
|
Control of glucose utilization in working perfused rat heart
|
|
3481
|
Control of glucose utilization in working perfused rat heart
|
|
3482
|
Control of glucose utilization in working perfused rat heart
|
|
3483
|
Control of glucose utilization in working perfused rat heart
|
|
3484
|
Control of glucose utilization in working perfused rat heart
|
|
3485
|
Control of glucose utilization in working perfused rat heart
|
|
3486
|
Control of glucose utilization in working perfused rat heart
|
|
3487
|
Control of glucose utilization in working perfused rat heart
|
|
3488
|
Control of glucose utilization in working perfused rat heart
|
|
3489
|
Control of glucose utilization in working perfused rat heart
|
|
3490
|
Control of glucose utilization in working perfused rat heart
|
|
3491
|
Control of glucose utilization in working perfused rat heart
|
|
3492
|
Control of glucose utilization in working perfused rat heart
|
|
3493
|
Control of glucose utilization in working perfused rat heart
|
|
3494
|
Control of glucose utilization in working perfused rat heart
|
|
3495
|
Control of glucose utilization in working perfused rat heart
|
|
3496
|
Snapshots of catalysis: the structure of fructose-1,6-(bis)phosphate aldolase covalently bound to the ...
|
|
3497
|
Control of the reactivation kinetics of homodimeric triosephosphate isomerase from unfolded monomers
|
|
3498
|
Control of the reactivation kinetics of homodimeric triosephosphate isomerase from unfolded monomers
|
|
3499
|
Control of the reactivation kinetics of homodimeric triosephosphate isomerase from unfolded monomers
|
|
3500
|
Control of the reactivation kinetics of homodimeric triosephosphate isomerase from unfolded monomers
|
|
3501
|
Purification of the Type II and Type III isozymes of rat hexokinase, expressed in yeast.
|
|
3502
|
Purification of the Type II and Type III isozymes of rat hexokinase, expressed in yeast.
|
|
3510
|
Control of dolichyl phosphoglucose formation in human liver microsomes. Kinetic and inhibition studies of ...
|
|
3511
|
Control of dolichyl phosphoglucose formation in human liver microsomes. Kinetic and inhibition studies of ...
|
|
3512
|
Characterization of a glucose-repressed pyruvate kinase (Pyk2p) in Saccharomyces cerevisiae that is ...
|
|
3513
|
Characterization of a glucose-repressed pyruvate kinase (Pyk2p) in Saccharomyces cerevisiae that is ...
|
|
3514
|
Characterization of a glucose-repressed pyruvate kinase (Pyk2p) in Saccharomyces cerevisiae that is ...
|
|
3515
|
Characterization of a glucose-repressed pyruvate kinase (Pyk2p) in Saccharomyces cerevisiae that is ...
|
|
3516
|
Characterization of a glucose-repressed pyruvate kinase (Pyk2p) in Saccharomyces cerevisiae that is ...
|
|
3517
|
Characterization of a glucose-repressed pyruvate kinase (Pyk2p) in Saccharomyces cerevisiae that is ...
|
|
3518
|
Kinetic studies of sheep kidney gamma-glutamyl transpeptidase
|
|
3519
|
Kinetic studies of sheep kidney gamma-glutamyl transpeptidase
|
|
3520
|
Kinetic studies of sheep kidney gamma-glutamyl transpeptidase
|
|
3521
|
YMXM motifs of IRS-1 define substrate specificity of the insulin receptor kinase
|
|
3522
|
YMXM motifs of IRS-1 define substrate specificity of the insulin receptor kinase
|
|
3523
|
YMXM motifs of IRS-1 define substrate specificity of the insulin receptor kinase
|
|
3524
|
YMXM motifs of IRS-1 define substrate specificity of the insulin receptor kinase
|
|
3525
|
YMXM motifs of IRS-1 define substrate specificity of the insulin receptor kinase
|
|
3526
|
YMXM motifs of IRS-1 define substrate specificity of the insulin receptor kinase
|
|
3527
|
YMXM motifs of IRS-1 define substrate specificity of the insulin receptor kinase
|
|
3528
|
YMXM motifs of IRS-1 define substrate specificity of the insulin receptor kinase
|
|
3529
|
YMXM motifs of IRS-1 define substrate specificity of the insulin receptor kinase
|
|
3530
|
YMXM motifs of IRS-1 define substrate specificity of the insulin receptor kinase
|
|
3531
|
YMXM motifs of IRS-1 define substrate specificity of the insulin receptor kinase
|
|
3532
|
YMXM motifs of IRS-1 define substrate specificity of the insulin receptor kinase
|
|
3533
|
YMXM motifs of IRS-1 define substrate specificity of the insulin receptor kinase
|
|
3534
|
YMXM motifs of IRS-1 define substrate specificity of the insulin receptor kinase
|
|
3535
|
YMXM motifs of IRS-1 define substrate specificity of the insulin receptor kinase
|
|
3536
|
Mechanistic insight into 3-deoxy-D-manno-octulosonate-8-phosphate synthase and ...
|
|
3537
|
Mechanistic insight into 3-deoxy-D-manno-octulosonate-8-phosphate synthase and ...
|
|
3538
|
Mechanistic insight into 3-deoxy-D-manno-octulosonate-8-phosphate synthase and ...
|
|
3539
|
Mechanistic insight into 3-deoxy-D-manno-octulosonate-8-phosphate synthase and ...
|
|
3544
|
Purification and Characterization of Phenylalanine 4-Monooxygenase from Rat Liver
|
|
3545
|
Purification and Characterization of Phenylalanine 4-Monooxygenase from Rat Liver
|
|
3546
|
Purification and Characterization of Phenylalanine 4-Monooxygenase from Rat Liver
|
|
3547
|
Purification and Characterization of Phenylalanine 4-Monooxygenase from Rat Liver
|
|
3548
|
Purification and Characterization of Phenylalanine 4-Monooxygenase from Rat Liver
|
|
3549
|
Purification and Characterization of Phenylalanine 4-Monooxygenase from Rat Liver
|
|
3550
|
Purification and Characterization of Phenylalanine 4-Monooxygenase from Rat Liver
|
|
3551
|
Purification and Characterization of Phenylalanine 4-Monooxygenase from Rat Liver
|
|
3552
|
Purification and Characterization of Phenylalanine 4-Monooxygenase from Rat Liver
|
|
3553
|
Purification and Characterization of Phenylalanine 4-Monooxygenase from Rat Liver
|
|
3554
|
Purification and Characterization of Phenylalanine 4-Monooxygenase from Rat Liver
|
|
3555
|
Purification and Characterization of Phenylalanine 4-Monooxygenase from Rat Liver
|
|
3556
|
Isolation and Characterization of a Novel Eukaryotic Monofunctional NAD+-Dependent ...
|
|
3557
|
Regulation of liver metabolism by enzyme phosphorylation during mammalian hibernation
|
|
3558
|
Regulation of liver metabolism by enzyme phosphorylation during mammalian hibernation
|
|
3559
|
Regulation of liver metabolism by enzyme phosphorylation during mammalian hibernation
|
|
3560
|
Regulation of liver metabolism by enzyme phosphorylation during mammalian hibernation
|
|
3561
|
Regulation of liver metabolism by enzyme phosphorylation during mammalian hibernation
|
|
3562
|
Regulation of liver metabolism by enzyme phosphorylation during mammalian hibernation
|
|
3563
|
Regulation of liver metabolism by enzyme phosphorylation during mammalian hibernation
|
|
3564
|
Regulation of liver metabolism by enzyme phosphorylation during mammalian hibernation
|
|
3565
|
Regulation of liver metabolism by enzyme phosphorylation during mammalian hibernation
|
|
3566
|
Regulation of liver metabolism by enzyme phosphorylation during mammalian hibernation
|
|
3567
|
Regulation of liver metabolism by enzyme phosphorylation during mammalian hibernation
|
|
3568
|
Regulation of liver metabolism by enzyme phosphorylation during mammalian hibernation
|
|
3569
|
Regulation of liver metabolism by enzyme phosphorylation during mammalian hibernation
|
|
3570
|
Regulation of liver metabolism by enzyme phosphorylation during mammalian hibernation
|
|
3571
|
Regulation of liver metabolism by enzyme phosphorylation during mammalian hibernation
|
|
3572
|
Regulation of liver metabolism by enzyme phosphorylation during mammalian hibernation
|
|
3573
|
Regulation of liver metabolism by enzyme phosphorylation during mammalian hibernation
|
|
3574
|
Regulation of liver metabolism by enzyme phosphorylation during mammalian hibernation
|
|
3575
|
Regulation of liver metabolism by enzyme phosphorylation during mammalian hibernation
|
|
3576
|
Regulation of liver metabolism by enzyme phosphorylation during mammalian hibernation
|
|
3577
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3578
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3579
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3580
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3581
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3582
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3583
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3584
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3585
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3586
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3587
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3588
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3589
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3590
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3591
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3592
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3593
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3594
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3595
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3596
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3597
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3598
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3599
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3600
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3601
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3602
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3603
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3604
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3605
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3606
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3607
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3608
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3609
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3610
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3611
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3612
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3613
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3614
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3615
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3616
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3617
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3618
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3619
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3620
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3621
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3622
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3623
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3624
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3625
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3626
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3627
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3628
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3629
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3630
|
The inhibitory effect of halides and carboxylates an hepatic NADH:cytochrome b5 oxidoreductase
|
|
3685
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3686
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3687
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3688
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3689
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3690
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3691
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3692
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3693
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3694
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3695
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3696
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3697
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3698
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3699
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3700
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3701
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3702
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3703
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3704
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3705
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3706
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3707
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3708
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3709
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3710
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3711
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3712
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3713
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3714
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3715
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3716
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3717
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3718
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3719
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3720
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3721
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3722
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3723
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3724
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3725
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3726
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3727
|
Functionally important amino acids in Saccharomyces cerevisiae aspartate kinase
|
|
3728
|
Cathepsin V, a novel and potent elastolytic activity expressed in activated macrophages
|
|
3729
|
Cathepsin V, a novel and potent elastolytic activity expressed in activated macrophages
|
|
3730
|
Cathepsin V, a novel and potent elastolytic activity expressed in activated macrophages
|
|
3731
|
Cathepsin V, a novel and potent elastolytic activity expressed in activated macrophages
|
|
3732
|
Kinetic characterization of phenol and aniline derivates as substrates of peroxidase
|
|
3733
|
Kinetic characterization of phenol and aniline derivates as substrates of peroxidase
|
|
3734
|
Kinetic characterization of phenol and aniline derivates as substrates of peroxidase
|
|
3735
|
Kinetic characterization of phenol and aniline derivates as substrates of peroxidase
|
|
3736
|
Kinetic characterization of phenol and aniline derivates as substrates of peroxidase
|
|
3737
|
Kinetic characterization of phenol and aniline derivates as substrates of peroxidase
|
|
3738
|
Kinetic characterization of phenol and aniline derivates as substrates of peroxidase
|
|
3739
|
Kinetic characterization of phenol and aniline derivates as substrates of peroxidase
|
|
3740
|
Kinetic characterization of phenol and aniline derivates as substrates of peroxidase
|
|
3741
|
Kinetic behavior of Desulfovibrio gigas aldehyde oxidoreductase encapsulated in reverse micelles
|
|
3742
|
Kinetic behavior of Desulfovibrio gigas aldehyde oxidoreductase encapsulated in reverse micelles
|
|
3743
|
Kinetic behavior of Desulfovibrio gigas aldehyde oxidoreductase encapsulated in reverse micelles
|
|
3744
|
Kinetic behavior of Desulfovibrio gigas aldehyde oxidoreductase encapsulated in reverse micelles
|
|
3745
|
Kinetic behavior of Desulfovibrio gigas aldehyde oxidoreductase encapsulated in reverse micelles
|
|
3746
|
Kinetic behavior of Desulfovibrio gigas aldehyde oxidoreductase encapsulated in reverse micelles
|
|
3747
|
Kinetic behavior of Desulfovibrio gigas aldehyde oxidoreductase encapsulated in reverse micelles
|
|
3748
|
A large hinge bending domain rotation is necessary for the catalytic function of Escherichia coli ...
|
|
3749
|
A large hinge bending domain rotation is necessary for the catalytic function of Escherichia coli ...
|
|
3750
|
A large hinge bending domain rotation is necessary for the catalytic function of Escherichia coli ...
|
|
3751
|
A large hinge bending domain rotation is necessary for the catalytic function of Escherichia coli ...
|
|
3752
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3753
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3754
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3755
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3756
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3757
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3758
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3759
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3760
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3761
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3762
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3763
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3764
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3765
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3766
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3767
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3768
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3769
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3770
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3771
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3772
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3773
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3774
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3775
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3776
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3777
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3778
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3779
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3780
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3781
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3782
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
|
|
3783
|
Novel psychrophilic and thermolabile L-threonine dehydrogenase from psychrophilic Cytophaga sp. strain KUC-1
|
|
3784
|
Novel psychrophilic and thermolabile L-threonine dehydrogenase from psychrophilic Cytophaga sp. strain KUC-1
|
|
3785
|
Novel psychrophilic and thermolabile L-threonine dehydrogenase from psychrophilic Cytophaga sp. strain KUC-1
|
|
3786
|
Novel psychrophilic and thermolabile L-threonine dehydrogenase from psychrophilic Cytophaga sp. strain KUC-1
|
|
3787
|
Novel psychrophilic and thermolabile L-threonine dehydrogenase from psychrophilic Cytophaga sp. strain KUC-1
|
|
3788
|
Novel psychrophilic and thermolabile L-threonine dehydrogenase from psychrophilic Cytophaga sp. strain KUC-1
|
|
3789
|
Novel psychrophilic and thermolabile L-threonine dehydrogenase from psychrophilic Cytophaga sp. strain KUC-1
|
|
3790
|
Novel psychrophilic and thermolabile L-threonine dehydrogenase from psychrophilic Cytophaga sp. strain KUC-1
|
|
3791
|
Novel psychrophilic and thermolabile L-threonine dehydrogenase from psychrophilic Cytophaga sp. strain KUC-1
|
|
3792
|
L-Threonine dehydrogenase of chicken liver. Purification, characterization, and physiological significance
|
|
3793
|
Multiple hydrogen kinetic isotope effects for enzymes catalyzing exchange with solvent: application to alanine ...
|
|
3794
|
Multiple hydrogen kinetic isotope effects for enzymes catalyzing exchange with solvent: application to alanine ...
|
|
3795
|
Multiple hydrogen kinetic isotope effects for enzymes catalyzing exchange with solvent: application to alanine ...
|
|
3796
|
Multiple hydrogen kinetic isotope effects for enzymes catalyzing exchange with solvent: application to alanine ...
|
|
3797
|
Crystal structure of human glutathione S-transferase A3-3 and mechanistic implications for its high steroid ...
|
|
3798
|
Crystal structure of human glutathione S-transferase A3-3 and mechanistic implications for its high steroid ...
|
|
3799
|
Crystal structure of human glutathione S-transferase A3-3 and mechanistic implications for its high steroid ...
|
|
3800
|
Crystal structure of human glutathione S-transferase A3-3 and mechanistic implications for its high steroid ...
|
|
3801
|
Effects of cryostabilizers, low temperature, and freezing on the kinetics of the pectin ...
|
|
3802
|
Effects of cryostabilizers, low temperature, and freezing on the kinetics of the pectin ...
|
|
3803
|
Effects of cryostabilizers, low temperature, and freezing on the kinetics of the pectin ...
|
|
3804
|
Organ-specific human alcohol dehydrogenase: Isolation and characterization of isozymes from testis
|
|
3805
|
Organ-specific human alcohol dehydrogenase: Isolation and characterization of isozymes from testis
|
|
3806
|
Organ-specific human alcohol dehydrogenase: Isolation and characterization of isozymes from testis
|
|
3807
|
Carboxymethylproline synthase from Pectobacterium carotorova: a multifaceted member of the crotonase ...
|
|
3808
|
Carboxymethylproline synthase from Pectobacterium carotorova: a multifaceted member of the crotonase ...
|
|
3809
|
Carboxymethylproline synthase from Pectobacterium carotorova: a multifaceted member of the crotonase ...
|
|
3810
|
Carboxymethylproline synthase from Pectobacterium carotorova: a multifaceted member of the crotonase ...
|
|
3811
|
Carboxymethylproline synthase from Pectobacterium carotorova: a multifaceted member of the crotonase ...
|
|
3812
|
Carboxymethylproline synthase from Pectobacterium carotorova: a multifaceted member of the crotonase ...
|
|
3813
|
Carboxymethylproline synthase from Pectobacterium carotorova: a multifaceted member of the crotonase ...
|
|
3814
|
Ornithine decarboxylase promotes catalysis by binding the carboxylate in a buried pocket containing ...
|
|
3815
|
The active site and mechanism of action of recombinant acetohydroxy acid synthase from tobacco
|
|
3816
|
The active site and mechanism of action of recombinant acetohydroxy acid synthase from tobacco
|
|
3817
|
Coenzyme site-directed mutants of photosynthetic A4-GAPDH show selectively reduced NADPH-dependent catalysis, ...
|
|
3818
|
Coenzyme site-directed mutants of photosynthetic A4-GAPDH show selectively reduced NADPH-dependent catalysis, ...
|
|
3819
|
Coenzyme site-directed mutants of photosynthetic A4-GAPDH show selectively reduced NADPH-dependent catalysis, ...
|
|
3820
|
Coenzyme site-directed mutants of photosynthetic A4-GAPDH show selectively reduced NADPH-dependent catalysis, ...
|
|
3821
|
Coenzyme site-directed mutants of photosynthetic A4-GAPDH show selectively reduced NADPH-dependent catalysis, ...
|
|
3822
|
Coenzyme site-directed mutants of photosynthetic A4-GAPDH show selectively reduced NADPH-dependent catalysis, ...
|
|
3823
|
Coenzyme site-directed mutants of photosynthetic A4-GAPDH show selectively reduced NADPH-dependent catalysis, ...
|
|
3824
|
Coenzyme site-directed mutants of photosynthetic A4-GAPDH show selectively reduced NADPH-dependent catalysis, ...
|
|
3825
|
Coenzyme site-directed mutants of photosynthetic A4-GAPDH show selectively reduced NADPH-dependent catalysis, ...
|
|
3826
|
Coenzyme site-directed mutants of photosynthetic A4-GAPDH show selectively reduced NADPH-dependent catalysis, ...
|
|
3827
|
Mouse liver CYP2C39 is a novel retinoic acid 4-hydroxylase. Its down-regulation offers a molecular basis for ...
|
|
3828
|
Production and characterization of the Talaromyces stipitatus feruloyl esterase FAEC in Pichia pastoris: ...
|
|
3829
|
Production and characterization of the Talaromyces stipitatus feruloyl esterase FAEC in Pichia pastoris: ...
|
|
3830
|
Production and characterization of the Talaromyces stipitatus feruloyl esterase FAEC in Pichia pastoris: ...
|
|
3831
|
Production and characterization of the Talaromyces stipitatus feruloyl esterase FAEC in Pichia pastoris: ...
|
|
3832
|
NAD-dependent methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase is the mammalian ...
|
|
3833
|
NAD-dependent methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase is the mammalian ...
|
|
3834
|
NAD-dependent methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase is the mammalian ...
|
|
3835
|
NAD-dependent methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase is the mammalian ...
|
|
3836
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3837
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3838
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3839
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3840
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3841
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3842
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3843
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3844
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3845
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3846
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3847
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3848
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3849
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3850
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3851
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3852
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3853
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3854
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3855
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3856
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3857
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3858
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3859
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3860
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3861
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3862
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3863
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3864
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3865
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3866
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3867
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3868
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3869
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3870
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3871
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3872
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3873
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3874
|
Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between ...
|
|
3875
|
Serine acetyltransferase of Escherichia coli: substrate specificity and feedback control by cysteine
|
|
3876
|
Serine acetyltransferase of Escherichia coli: substrate specificity and feedback control by cysteine
|
|
3877
|
Serine acetyltransferase of Escherichia coli: substrate specificity and feedback control by cysteine
|
|
3878
|
Serine acetyltransferase of Escherichia coli: substrate specificity and feedback control by cysteine
|
|
3879
|
Serine acetyltransferase of Escherichia coli: substrate specificity and feedback control by cysteine
|
|
3880
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3881
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3882
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3883
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3884
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3885
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3886
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3887
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3888
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3889
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3890
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3891
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3892
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3893
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3894
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3895
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3896
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3897
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3898
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3899
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3900
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3901
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3902
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3903
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3904
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3905
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3906
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3907
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3908
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3909
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3910
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3911
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3912
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3913
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3914
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3915
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3916
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3917
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3918
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3919
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3920
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3921
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3922
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3923
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3924
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3925
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3926
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3927
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3928
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3929
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3930
|
Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B
|
|
3931
|
Maize C4 NADP-malic enzyme. Expression in Escherichia coli and characterization of site-directed mutants at ...
|
|
3932
|
Maize C4 NADP-malic enzyme. Expression in Escherichia coli and characterization of site-directed mutants at ...
|
|
3933
|
Maize C4 NADP-malic enzyme. Expression in Escherichia coli and characterization of site-directed mutants at ...
|
|
3934
|
Maize C4 NADP-malic enzyme. Expression in Escherichia coli and characterization of site-directed mutants at ...
|
|
3935
|
Maize C4 NADP-malic enzyme. Expression in Escherichia coli and characterization of site-directed mutants at ...
|
|
3936
|
Maize C4 NADP-malic enzyme. Expression in Escherichia coli and characterization of site-directed mutants at ...
|
|
3937
|
Maize C4 NADP-malic enzyme. Expression in Escherichia coli and characterization of site-directed mutants at ...
|
|
3938
|
Structural and functional impairment of an Old Yellow Enzyme homologue upon affinity tag incorporation
|
|
3939
|
Structural and functional impairment of an Old Yellow Enzyme homologue upon affinity tag incorporation
|
|
3940
|
Structural and functional impairment of an Old Yellow Enzyme homologue upon affinity tag incorporation
|
|
3941
|
Structural and functional impairment of an Old Yellow Enzyme homologue upon affinity tag incorporation
|
|
3942
|
Structural and functional impairment of an Old Yellow Enzyme homologue upon affinity tag incorporation
|
|
3943
|
Chromogenic assay for the activity of sphingomyelinase from Bacillus cereus and its application to the ...
|
|
3944
|
Chromogenic assay for the activity of sphingomyelinase from Bacillus cereus and its application to the ...
|
|
3945
|
Chromogenic assay for the activity of sphingomyelinase from Bacillus cereus and its application to the ...
|
|
3946
|
Chromogenic assay for the activity of sphingomyelinase from Bacillus cereus and its application to the ...
|
|
3947
|
Chromogenic assay for the activity of sphingomyelinase from Bacillus cereus and its application to the ...
|
|
3948
|
Chromogenic assay for the activity of sphingomyelinase from Bacillus cereus and its application to the ...
|
|
3949
|
Chromogenic assay for the activity of sphingomyelinase from Bacillus cereus and its application to the ...
|
|
3950
|
Chromogenic assay for the activity of sphingomyelinase from Bacillus cereus and its application to the ...
|
|
3951
|
Chromogenic assay for the activity of sphingomyelinase from Bacillus cereus and its application to the ...
|
|
3952
|
Reactions of trans-3-chloroacrylic acid dehalogenase with acetylene substrates: consequences of and evidence ...
|
|
3953
|
Reactions of trans-3-chloroacrylic acid dehalogenase with acetylene substrates: consequences of and evidence ...
|
|
3954
|
Reactions of trans-3-chloroacrylic acid dehalogenase with acetylene substrates: consequences of and evidence ...
|
|
3955
|
Spatial activities and induction of glutamate-cysteine ligase (GCL) in the postimplantation rat embryo and ...
|
|
3956
|
Spatial activities and induction of glutamate-cysteine ligase (GCL) in the postimplantation rat embryo and ...
|
|
3957
|
Spatial activities and induction of glutamate-cysteine ligase (GCL) in the postimplantation rat embryo and ...
|
|
3958
|
Spatial activities and induction of glutamate-cysteine ligase (GCL) in the postimplantation rat embryo and ...
|
|
3959
|
Kinetic analysis of Escherichia coli 2-C-methyl-D-erythritol-4-phosphate cytidyltransferase, wild type and ...
|
|
3960
|
Kinetic analysis of Escherichia coli 2-C-methyl-D-erythritol-4-phosphate cytidyltransferase, wild type and ...
|
|
3961
|
Kinetic analysis of Escherichia coli 2-C-methyl-D-erythritol-4-phosphate cytidyltransferase, wild type and ...
|
|
3962
|
Kinetic analysis of Escherichia coli 2-C-methyl-D-erythritol-4-phosphate cytidyltransferase, wild type and ...
|
|
3963
|
Kinetic analysis of Escherichia coli 2-C-methyl-D-erythritol-4-phosphate cytidyltransferase, wild type and ...
|
|
3964
|
Kinetic analysis of Escherichia coli 2-C-methyl-D-erythritol-4-phosphate cytidyltransferase, wild type and ...
|
|
3965
|
Kinetic analysis of Escherichia coli 2-C-methyl-D-erythritol-4-phosphate cytidyltransferase, wild type and ...
|
|
3966
|
Kinetic analysis of Escherichia coli 2-C-methyl-D-erythritol-4-phosphate cytidyltransferase, wild type and ...
|
|
3967
|
Kinetic analysis of Escherichia coli 2-C-methyl-D-erythritol-4-phosphate cytidyltransferase, wild type and ...
|
|
3968
|
Kinetic analysis of Escherichia coli 2-C-methyl-D-erythritol-4-phosphate cytidyltransferase, wild type and ...
|
|
3969
|
Kinetic analysis of Escherichia coli 2-C-methyl-D-erythritol-4-phosphate cytidyltransferase, wild type and ...
|
|
3970
|
Kinetic analysis of Escherichia coli 2-C-methyl-D-erythritol-4-phosphate cytidyltransferase, wild type and ...
|
|
3971
|
Kinetic analysis of Escherichia coli 2-C-methyl-D-erythritol-4-phosphate cytidyltransferase, wild type and ...
|
|
3972
|
Kinetic analysis of Escherichia coli 2-C-methyl-D-erythritol-4-phosphate cytidyltransferase, wild type and ...
|
|
3973
|
Kinetic analysis of Escherichia coli 2-C-methyl-D-erythritol-4-phosphate cytidyltransferase, wild type and ...
|
|
3974
|
Kinetic analysis of Escherichia coli 2-C-methyl-D-erythritol-4-phosphate cytidyltransferase, wild type and ...
|
|
3975
|
Kinetic analysis of Escherichia coli 2-C-methyl-D-erythritol-4-phosphate cytidyltransferase, wild type and ...
|
|
3976
|
Kinetic analysis of Escherichia coli 2-C-methyl-D-erythritol-4-phosphate cytidyltransferase, wild type and ...
|
|
3977
|
A kinetic analysis of the nucleotide-induced allosteric transitions in a single-ring mutant of GroEL
|
|
3978
|
A kinetic analysis of the nucleotide-induced allosteric transitions in a single-ring mutant of GroEL
|
|
3979
|
A kinetic analysis of the nucleotide-induced allosteric transitions in a single-ring mutant of GroEL
|
|
3980
|
Asparagine 285 plays a key role in transition state stabilization in rabbit muscle creatine kinase
|
|
3981
|
Asparagine 285 plays a key role in transition state stabilization in rabbit muscle creatine kinase
|
|
3982
|
Asparagine 285 plays a key role in transition state stabilization in rabbit muscle creatine kinase
|
|
3983
|
Asparagine 285 plays a key role in transition state stabilization in rabbit muscle creatine kinase
|
|
3984
|
Asparagine 285 plays a key role in transition state stabilization in rabbit muscle creatine kinase
|
|
3985
|
Asparagine 285 plays a key role in transition state stabilization in rabbit muscle creatine kinase
|
|
3986
|
Asparagine 285 plays a key role in transition state stabilization in rabbit muscle creatine kinase
|
|
3987
|
Asparagine 285 plays a key role in transition state stabilization in rabbit muscle creatine kinase
|
|
3988
|
Asparagine 285 plays a key role in transition state stabilization in rabbit muscle creatine kinase
|
|
3989
|
Asparagine 285 plays a key role in transition state stabilization in rabbit muscle creatine kinase
|
|
3990
|
Asparagine 285 plays a key role in transition state stabilization in rabbit muscle creatine kinase
|
|
3991
|
Asparagine 285 plays a key role in transition state stabilization in rabbit muscle creatine kinase
|
|
3992
|
Asparagine 285 plays a key role in transition state stabilization in rabbit muscle creatine kinase
|
|
3993
|
Asparagine 285 plays a key role in transition state stabilization in rabbit muscle creatine kinase
|
|
3994
|
Asparagine 285 plays a key role in transition state stabilization in rabbit muscle creatine kinase
|
|
3995
|
Asparagine 285 plays a key role in transition state stabilization in rabbit muscle creatine kinase
|
|
3996
|
Asparagine 285 plays a key role in transition state stabilization in rabbit muscle creatine kinase
|
|
3997
|
Asparagine 285 plays a key role in transition state stabilization in rabbit muscle creatine kinase
|
|
3998
|
Asparagine 285 plays a key role in transition state stabilization in rabbit muscle creatine kinase
|
|
3999
|
Effects of multiple ligand binding on kinetic isotope effects in PQQ-dependent methanol dehydrogenase
|
|
4000
|
Effects of multiple ligand binding on kinetic isotope effects in PQQ-dependent methanol dehydrogenase
|





 If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.
If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.