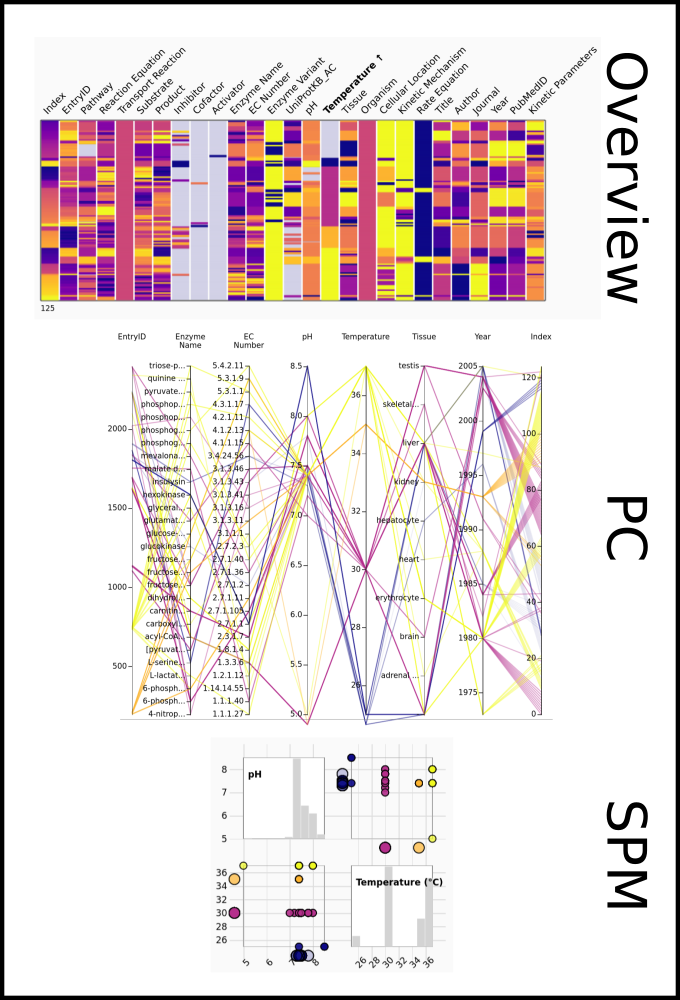
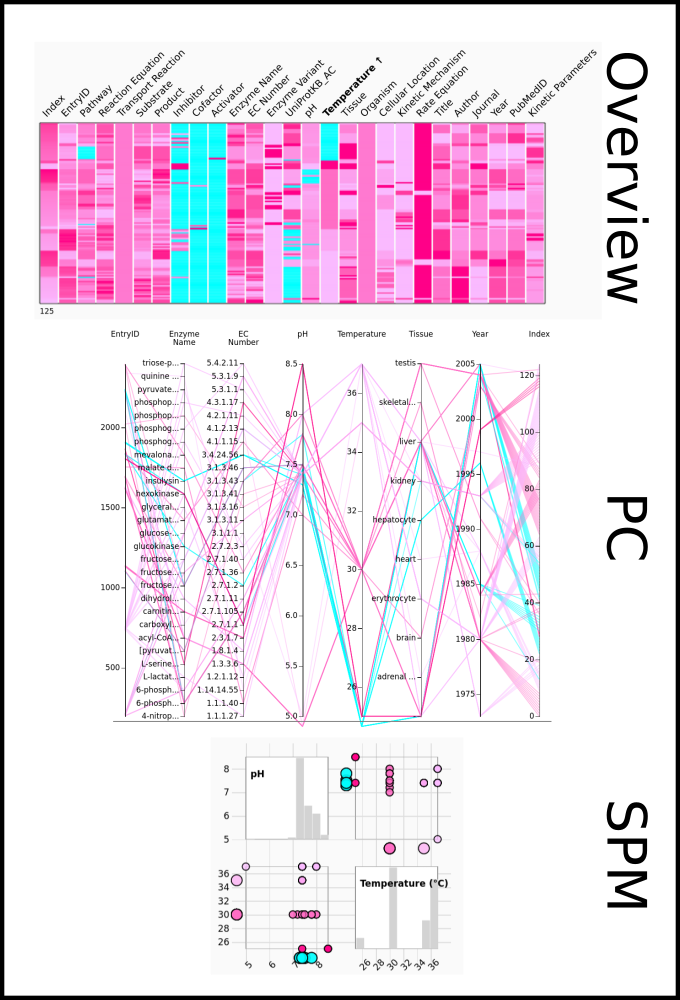
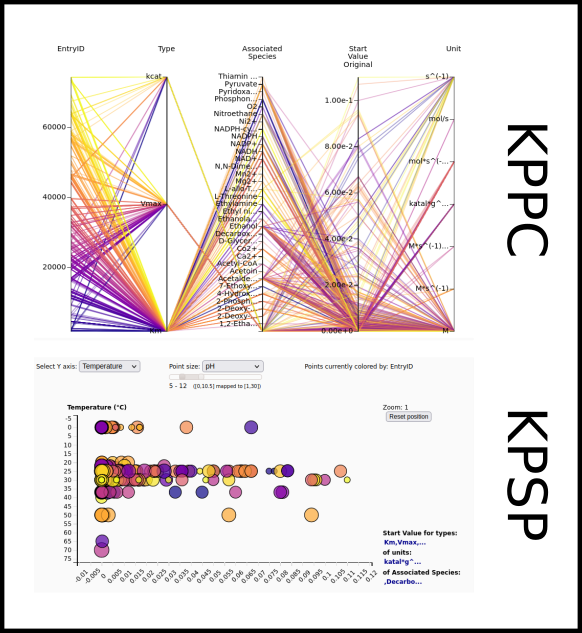
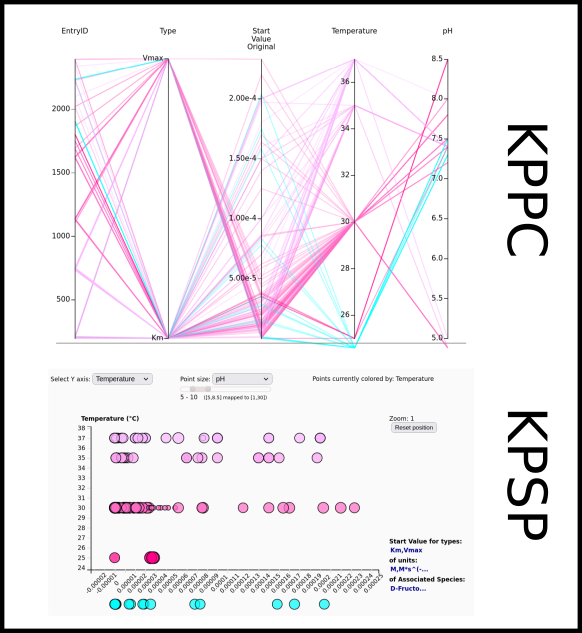
To improve SABIO-RK data interoperability, semantic markup was added to web pages as described and defined by
This structured information makes it easier to discover, collate and analyse our data.
|
25001
|
Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from ...
|
|
25002
|
Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from ...
|
|
25003
|
Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from ...
|
|
25004
|
Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from ...
|
|
25005
|
Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from ...
|
|
25006
|
Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from ...
|
|
25007
|
Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from ...
|
|
25008
|
Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from ...
|
|
25009
|
Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from ...
|
|
25010
|
Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from ...
|
|
25011
|
Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from ...
|
|
25012
|
Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from ...
|
|
25013
|
Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from ...
|
|
25014
|
Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from ...
|
|
25015
|
Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from ...
|
|
25016
|
Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from ...
|
|
25017
|
Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from ...
|
|
25018
|
Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from ...
|
|
25019
|
Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from ...
|
|
25020
|
Purification and properties of the enzyme 6-phosphogluconate dehydrogenase from Dicentrarchus labrax L. liver
|
|
25021
|
Purification and properties of the enzyme 6-phosphogluconate dehydrogenase from Dicentrarchus labrax L. liver
|
|
25022
|
Purification and properties of the enzyme 6-phosphogluconate dehydrogenase from Dicentrarchus labrax L. liver
|
|
25023
|
Purification and properties of the enzyme 6-phosphogluconate dehydrogenase from Dicentrarchus labrax L. liver
|
|
25024
|
Purification and properties of the enzyme 6-phosphogluconate dehydrogenase from Dicentrarchus labrax L. liver
|
|
25025
|
Identification of 7 alpha,12 alpha-dihydroxy-5 beta-cholestan-3-one 3 alpha-hydroxysteroid dehydrogenase
|
|
25026
|
Identification of 7 alpha,12 alpha-dihydroxy-5 beta-cholestan-3-one 3 alpha-hydroxysteroid dehydrogenase
|
|
25027
|
Identification of 7 alpha,12 alpha-dihydroxy-5 beta-cholestan-3-one 3 alpha-hydroxysteroid dehydrogenase
|
|
25028
|
Identification of 7 alpha,12 alpha-dihydroxy-5 beta-cholestan-3-one 3 alpha-hydroxysteroid dehydrogenase
|
|
25029
|
Identification of 7 alpha,12 alpha-dihydroxy-5 beta-cholestan-3-one 3 alpha-hydroxysteroid dehydrogenase
|
|
25030
|
Identification of 7 alpha,12 alpha-dihydroxy-5 beta-cholestan-3-one 3 alpha-hydroxysteroid dehydrogenase
|
|
25031
|
Identification of 7 alpha,12 alpha-dihydroxy-5 beta-cholestan-3-one 3 alpha-hydroxysteroid dehydrogenase
|
|
25032
|
Identification of 7 alpha,12 alpha-dihydroxy-5 beta-cholestan-3-one 3 alpha-hydroxysteroid dehydrogenase
|
|
25033
|
Identification of 7 alpha,12 alpha-dihydroxy-5 beta-cholestan-3-one 3 alpha-hydroxysteroid dehydrogenase
|
|
25034
|
Identification of 7 alpha,12 alpha-dihydroxy-5 beta-cholestan-3-one 3 alpha-hydroxysteroid dehydrogenase
|
|
25035
|
Identification of 7 alpha,12 alpha-dihydroxy-5 beta-cholestan-3-one 3 alpha-hydroxysteroid dehydrogenase
|
|
25036
|
Identification of 7 alpha,12 alpha-dihydroxy-5 beta-cholestan-3-one 3 alpha-hydroxysteroid dehydrogenase
|
|
25037
|
Identification of 7 alpha,12 alpha-dihydroxy-5 beta-cholestan-3-one 3 alpha-hydroxysteroid dehydrogenase
|
|
25038
|
Identification of 7 alpha,12 alpha-dihydroxy-5 beta-cholestan-3-one 3 alpha-hydroxysteroid dehydrogenase
|
|
25039
|
Identification of 7 alpha,12 alpha-dihydroxy-5 beta-cholestan-3-one 3 alpha-hydroxysteroid dehydrogenase
|
|
25040
|
Functional Consequences and Exonuclease Kinetic Parameters of Point Mutations in Bacteriophage T4 DNA ...
|
|
25041
|
Functional Consequences and Exonuclease Kinetic Parameters of Point Mutations in Bacteriophage T4 DNA ...
|
|
25042
|
Functional Consequences and Exonuclease Kinetic Parameters of Point Mutations in Bacteriophage T4 DNA ...
|
|
25043
|
Functional Consequences and Exonuclease Kinetic Parameters of Point Mutations in Bacteriophage T4 DNA ...
|
|
25044
|
Functional Consequences and Exonuclease Kinetic Parameters of Point Mutations in Bacteriophage T4 DNA ...
|
|
25045
|
Functional Consequences and Exonuclease Kinetic Parameters of Point Mutations in Bacteriophage T4 DNA ...
|
|
25046
|
Functional Consequences and Exonuclease Kinetic Parameters of Point Mutations in Bacteriophage T4 DNA ...
|
|
25047
|
Functional Consequences and Exonuclease Kinetic Parameters of Point Mutations in Bacteriophage T4 DNA ...
|
|
25048
|
Functional Consequences and Exonuclease Kinetic Parameters of Point Mutations in Bacteriophage T4 DNA ...
|
|
25049
|
Functional Consequences and Exonuclease Kinetic Parameters of Point Mutations in Bacteriophage T4 DNA ...
|
|
25050
|
Functional Consequences and Exonuclease Kinetic Parameters of Point Mutations in Bacteriophage T4 DNA ...
|
|
25051
|
Functional Consequences and Exonuclease Kinetic Parameters of Point Mutations in Bacteriophage T4 DNA ...
|
|
25052
|
The Comparative Enzymology of Lactic Dehydrogenases. I. Properties of the Crystalline Beef and Chicken Enzymes
|
|
25053
|
The Comparative Enzymology of Lactic Dehydrogenases. I. Properties of the Crystalline Beef and Chicken Enzymes
|
|
25054
|
The Comparative Enzymology of Lactic Dehydrogenases. I. Properties of the Crystalline Beef and Chicken Enzymes
|
|
25055
|
The Comparative Enzymology of Lactic Dehydrogenases. I. Properties of the Crystalline Beef and Chicken Enzymes
|
|
25056
|
The Comparative Enzymology of Lactic Dehydrogenases. I. Properties of the Crystalline Beef and Chicken Enzymes
|
|
25057
|
The Comparative Enzymology of Lactic Dehydrogenases. I. Properties of the Crystalline Beef and Chicken Enzymes
|
|
25058
|
The Comparative Enzymology of Lactic Dehydrogenases. I. Properties of the Crystalline Beef and Chicken Enzymes
|
|
25059
|
The Comparative Enzymology of Lactic Dehydrogenases. I. Properties of the Crystalline Beef and Chicken Enzymes
|
|
25060
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25061
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25062
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25063
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25064
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25065
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25066
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25067
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25068
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25069
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25070
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25071
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25072
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25073
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25074
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25075
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25076
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25077
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25078
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25079
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25080
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25081
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25082
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25083
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25084
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25085
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25086
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25087
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25088
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25089
|
Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo beta-lactamase
|
|
25090
|
Short and long term regulation of catecholamine biosynthetic enzymes by angiotensin in cultured adrenal ...
|
|
25091
|
Short and long term regulation of catecholamine biosynthetic enzymes by angiotensin in cultured adrenal ...
|
|
25092
|
Short and long term regulation of catecholamine biosynthetic enzymes by angiotensin in cultured adrenal ...
|
|
25093
|
alpha-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci
|
|
25094
|
Human brain short chain L-3-hydroxyacyl coenzyme A dehydrogenase is a single-domain multifunctional enzyme. ...
|
|
25095
|
Human brain short chain L-3-hydroxyacyl coenzyme A dehydrogenase is a single-domain multifunctional enzyme. ...
|
|
25096
|
Human brain short chain L-3-hydroxyacyl coenzyme A dehydrogenase is a single-domain multifunctional enzyme. ...
|
|
25097
|
Human brain short chain L-3-hydroxyacyl coenzyme A dehydrogenase is a single-domain multifunctional enzyme. ...
|
|
25098
|
Human brain short chain L-3-hydroxyacyl coenzyme A dehydrogenase is a single-domain multifunctional enzyme. ...
|
|
25099
|
Human brain short chain L-3-hydroxyacyl coenzyme A dehydrogenase is a single-domain multifunctional enzyme. ...
|
|
25100
|
Human brain short chain L-3-hydroxyacyl coenzyme A dehydrogenase is a single-domain multifunctional enzyme. ...
|
|
25101
|
Human brain short chain L-3-hydroxyacyl coenzyme A dehydrogenase is a single-domain multifunctional enzyme. ...
|
|
25102
|
Human brain short chain L-3-hydroxyacyl coenzyme A dehydrogenase is a single-domain multifunctional enzyme. ...
|
|
25103
|
Human brain short chain L-3-hydroxyacyl coenzyme A dehydrogenase is a single-domain multifunctional enzyme. ...
|
|
25104
|
Human brain short chain L-3-hydroxyacyl coenzyme A dehydrogenase is a single-domain multifunctional enzyme. ...
|
|
25105
|
Human brain short chain L-3-hydroxyacyl coenzyme A dehydrogenase is a single-domain multifunctional enzyme. ...
|
|
25106
|
Human brain short chain L-3-hydroxyacyl coenzyme A dehydrogenase is a single-domain multifunctional enzyme. ...
|
|
25107
|
Human brain short chain L-3-hydroxyacyl coenzyme A dehydrogenase is a single-domain multifunctional enzyme. ...
|
|
25108
|
Human brain short chain L-3-hydroxyacyl coenzyme A dehydrogenase is a single-domain multifunctional enzyme. ...
|
|
25109
|
Human brain short chain L-3-hydroxyacyl coenzyme A dehydrogenase is a single-domain multifunctional enzyme. ...
|
|
25110
|
D-Ribulose 5-phosphate 3-epimerase: functional and structural relationships to members of the ...
|
|
25111
|
D-Ribulose 5-phosphate 3-epimerase: functional and structural relationships to members of the ...
|
|
25112
|
D-Ribulose 5-phosphate 3-epimerase: functional and structural relationships to members of the ...
|
|
25113
|
D-Ribulose 5-phosphate 3-epimerase: functional and structural relationships to members of the ...
|
|
25114
|
D-Ribulose 5-phosphate 3-epimerase: functional and structural relationships to members of the ...
|
|
25115
|
D-Ribulose 5-phosphate 3-epimerase: functional and structural relationships to members of the ...
|
|
25116
|
D-Ribulose 5-phosphate 3-epimerase: functional and structural relationships to members of the ...
|
|
25117
|
Human liver cytosolic malate dehydrogenase: purification, kinetic properties, and role in ethanol metabolism
|
|
25118
|
Human liver cytosolic malate dehydrogenase: purification, kinetic properties, and role in ethanol metabolism
|
|
25119
|
Human liver cytosolic malate dehydrogenase: purification, kinetic properties, and role in ethanol metabolism
|
|
25120
|
Selenium-dependent and non-selenium-dependent glutathione peroxidases in human tissue extracts
|
|
25121
|
Selenium-dependent and non-selenium-dependent glutathione peroxidases in human tissue extracts
|
|
25122
|
Selenium-dependent and non-selenium-dependent glutathione peroxidases in human tissue extracts
|
|
25123
|
Human isopentenyl diphosphate: dimethylallyl diphosphate isomerase: overproduction, purification, and ...
|
|
25124
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25125
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25126
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25127
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25128
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25129
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25130
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25131
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25132
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25133
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25134
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25135
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25136
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25137
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25138
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25139
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25140
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25141
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25142
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25143
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25144
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25145
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25146
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25147
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25148
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25149
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25150
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25151
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25152
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25153
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25154
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25155
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25156
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25157
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25158
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25159
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25160
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25161
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25162
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25163
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25164
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25165
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25166
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25167
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25168
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25169
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25170
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25171
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25172
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25173
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25174
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25175
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25176
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25177
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25178
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25179
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25180
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25181
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25182
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25183
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal PepVs
|
|
25184
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25185
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25186
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25187
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25188
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25189
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25190
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25191
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25192
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25193
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25194
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25195
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25196
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25197
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25198
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25199
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25200
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25201
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25202
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25203
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25204
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25205
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25206
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25207
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25208
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25209
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25210
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25211
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25212
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25213
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25214
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25215
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25216
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25217
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25218
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25219
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25220
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25221
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25222
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25223
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25224
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25225
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25226
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25227
|
Structural elements in dextran glucosidase responsible for high specificity to long chain substrate
|
|
25228
|
Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes
|
|
25229
|
Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes
|
|
25230
|
Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes
|
|
25231
|
Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes
|
|
25232
|
Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes
|
|
25233
|
Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes
|
|
25234
|
Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes
|
|
25235
|
Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes
|
|
25236
|
Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes
|
|
25237
|
Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes
|
|
25238
|
Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes
|
|
25239
|
Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes
|
|
25240
|
Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes
|
|
25241
|
Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes
|
|
25242
|
Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes
|
|
25243
|
Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes
|
|
25244
|
Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes
|
|
25245
|
Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes
|
|
25246
|
Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes
|
|
25247
|
Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes
|
|
25248
|
Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes
|
|
25249
|
Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes
|
|
25250
|
Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes
|
|
25251
|
Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes
|
|
25252
|
Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes
|
|
25253
|
Horse liver aldehyde dehydrogenase. Purification and characterization of two isozymes
|
|
25254
|
Isolation and characterization of propionyl-CoA carboxylase from normal human liver. Evidence for a protomeric ...
|
|
25255
|
Isolation and characterization of propionyl-CoA carboxylase from normal human liver. Evidence for a protomeric ...
|
|
25256
|
Isolation and characterization of propionyl-CoA carboxylase from normal human liver. Evidence for a protomeric ...
|
|
25257
|
Isolation and characterization of propionyl-CoA carboxylase from normal human liver. Evidence for a protomeric ...
|
|
25258
|
The bifunctional active site of S-adenosylmethionine synthetase. Roles of the basic residues.
|
|
25259
|
The bifunctional active site of S-adenosylmethionine synthetase. Roles of the basic residues.
|
|
25260
|
The bifunctional active site of S-adenosylmethionine synthetase. Roles of the basic residues.
|
|
25261
|
The bifunctional active site of S-adenosylmethionine synthetase. Roles of the basic residues.
|
|
25262
|
The bifunctional active site of S-adenosylmethionine synthetase. Roles of the basic residues.
|
|
25263
|
The bifunctional active site of S-adenosylmethionine synthetase. Roles of the basic residues.
|
|
25264
|
The bifunctional active site of S-adenosylmethionine synthetase. Roles of the basic residues.
|
|
25265
|
The bifunctional active site of S-adenosylmethionine synthetase. Roles of the basic residues.
|
|
25266
|
The bifunctional active site of S-adenosylmethionine synthetase. Roles of the basic residues.
|
|
25267
|
The bifunctional active site of S-adenosylmethionine synthetase. Roles of the basic residues.
|
|
25268
|
The bifunctional active site of S-adenosylmethionine synthetase. Roles of the basic residues.
|
|
25269
|
The bifunctional active site of S-adenosylmethionine synthetase. Roles of the basic residues.
|
|
25270
|
The bifunctional active site of S-adenosylmethionine synthetase. Roles of the basic residues.
|
|
25271
|
The bifunctional active site of S-adenosylmethionine synthetase. Roles of the basic residues.
|
|
25272
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal ...
|
|
25273
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal ...
|
|
25274
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal ...
|
|
25275
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal ...
|
|
25276
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal ...
|
|
25277
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal ...
|
|
25278
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal ...
|
|
25279
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal ...
|
|
25280
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal ...
|
|
25281
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal ...
|
|
25282
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal ...
|
|
25283
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal ...
|
|
25284
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal ...
|
|
25285
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal ...
|
|
25286
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal ...
|
|
25287
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal ...
|
|
25288
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal ...
|
|
25289
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal ...
|
|
25290
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal ...
|
|
25291
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal ...
|
|
25292
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal ...
|
|
25293
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal ...
|
|
25294
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal ...
|
|
25295
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal ...
|
|
25296
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal ...
|
|
25297
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal ...
|
|
25298
|
Characterization and kinetic analysis of enzyme-substrate recognition by three recombinant lactococcal ...
|
|
25299
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25300
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25301
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25302
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25303
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25304
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25305
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25306
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25307
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25308
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25309
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25310
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25311
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25312
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25313
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25314
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25315
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25316
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25317
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25318
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25319
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25320
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25321
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25322
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25323
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25324
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25325
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25326
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25327
|
Regulation of deoxynucleoside kinase activities in rat liver mitochondria
|
|
25328
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25329
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25330
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25331
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25332
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25333
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25334
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25335
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25336
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25337
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25338
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25339
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25340
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25341
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25342
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25343
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25344
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25345
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25346
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25347
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25348
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25349
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25350
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25351
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25352
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25353
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25354
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25355
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25356
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25357
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25358
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25359
|
Substrate specificity in glycoside hydrolase family 10. Tyrosine 87 and leucine 314 play a pivotal role in ...
|
|
25360
|
Improved specificity toward substrates with positively charged side chains by site-directed mutagenesis of the ...
|
|
25361
|
Improved specificity toward substrates with positively charged side chains by site-directed mutagenesis of the ...
|
|
25362
|
Improved specificity toward substrates with positively charged side chains by site-directed mutagenesis of the ...
|
|
25363
|
Improved specificity toward substrates with positively charged side chains by site-directed mutagenesis of the ...
|
|
25364
|
Improved specificity toward substrates with positively charged side chains by site-directed mutagenesis of the ...
|
|
25365
|
Improved specificity toward substrates with positively charged side chains by site-directed mutagenesis of the ...
|
|
25366
|
Improved specificity toward substrates with positively charged side chains by site-directed mutagenesis of the ...
|
|
25367
|
Improved specificity toward substrates with positively charged side chains by site-directed mutagenesis of the ...
|
|
25368
|
Improved specificity toward substrates with positively charged side chains by site-directed mutagenesis of the ...
|
|
25369
|
Improved specificity toward substrates with positively charged side chains by site-directed mutagenesis of the ...
|
|
25370
|
Improved specificity toward substrates with positively charged side chains by site-directed mutagenesis of the ...
|
|
25371
|
Improved specificity toward substrates with positively charged side chains by site-directed mutagenesis of the ...
|
|
25372
|
Improved specificity toward substrates with positively charged side chains by site-directed mutagenesis of the ...
|
|
25373
|
Improved specificity toward substrates with positively charged side chains by site-directed mutagenesis of the ...
|
|
25374
|
Improved specificity toward substrates with positively charged side chains by site-directed mutagenesis of the ...
|
|
25375
|
Improved specificity toward substrates with positively charged side chains by site-directed mutagenesis of the ...
|
|
25376
|
Improved specificity toward substrates with positively charged side chains by site-directed mutagenesis of the ...
|
|
25377
|
Improved specificity toward substrates with positively charged side chains by site-directed mutagenesis of the ...
|
|
25378
|
Improved specificity toward substrates with positively charged side chains by site-directed mutagenesis of the ...
|
|
25379
|
Improved specificity toward substrates with positively charged side chains by site-directed mutagenesis of the ...
|
|
25380
|
Improved specificity toward substrates with positively charged side chains by site-directed mutagenesis of the ...
|
|
25381
|
Improved specificity toward substrates with positively charged side chains by site-directed mutagenesis of the ...
|
|
25382
|
Improved specificity toward substrates with positively charged side chains by site-directed mutagenesis of the ...
|
|
25383
|
Improved specificity toward substrates with positively charged side chains by site-directed mutagenesis of the ...
|
|
25384
|
Improved specificity toward substrates with positively charged side chains by site-directed mutagenesis of the ...
|
|
25385
|
Intragenic complementation and the structure and function of argininosuccinate lyase
|
|
25386
|
Intragenic complementation and the structure and function of argininosuccinate lyase
|
|
25387
|
Intragenic complementation and the structure and function of argininosuccinate lyase
|
|
25388
|
Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor
|
|
25389
|
Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor
|
|
25390
|
Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor
|
|
25391
|
Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor
|
|
25392
|
Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor
|
|
25393
|
Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor
|
|
25394
|
Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor
|
|
25395
|
Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor
|
|
25396
|
Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor
|
|
25397
|
Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor
|
|
25398
|
Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor
|
|
25399
|
Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor
|
|
25400
|
Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor
|
|
25401
|
Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor
|
|
25402
|
Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor
|
|
25403
|
Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor
|
|
25404
|
Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor
|
|
25405
|
Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor
|
|
25406
|
Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor
|
|
25407
|
Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor
|
|
25408
|
Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor
|
|
25409
|
Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor
|
|
25410
|
Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor
|
|
25411
|
Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor
|
|
25412
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25413
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25414
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25415
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25416
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25417
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25418
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25419
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25420
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25421
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25422
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25423
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25424
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25425
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25426
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25427
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25428
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25429
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25430
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25431
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25432
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25433
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25434
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25435
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25436
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25437
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25438
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25439
|
Identification of cysteine and arginine residues essential for the phosphotransacetylase from Methanosarcina ...
|
|
25440
|
Interactions of quinones with thioredoxin reductase: a challenge to the antioxidant role of the mammalian ...
|
|
25441
|
Interactions of quinones with thioredoxin reductase: a challenge to the antioxidant role of the mammalian ...
|
|
25442
|
Interactions of quinones with thioredoxin reductase: a challenge to the antioxidant role of the mammalian ...
|
|
25443
|
Interactions of quinones with thioredoxin reductase: a challenge to the antioxidant role of the mammalian ...
|
|
25444
|
Interactions of quinones with thioredoxin reductase: a challenge to the antioxidant role of the mammalian ...
|
|
25445
|
Interactions of quinones with thioredoxin reductase: a challenge to the antioxidant role of the mammalian ...
|
|
25446
|
Interactions of quinones with thioredoxin reductase: a challenge to the antioxidant role of the mammalian ...
|
|
25447
|
Interactions of quinones with thioredoxin reductase: a challenge to the antioxidant role of the mammalian ...
|
|
25448
|
Interactions of quinones with thioredoxin reductase: a challenge to the antioxidant role of the mammalian ...
|
|
25449
|
Interactions of quinones with thioredoxin reductase: a challenge to the antioxidant role of the mammalian ...
|
|
25450
|
Interactions of quinones with thioredoxin reductase: a challenge to the antioxidant role of the mammalian ...
|
|
25451
|
Interactions of quinones with thioredoxin reductase: a challenge to the antioxidant role of the mammalian ...
|
|
25452
|
Interactions of quinones with thioredoxin reductase: a challenge to the antioxidant role of the mammalian ...
|
|
25453
|
Interactions of quinones with thioredoxin reductase: a challenge to the antioxidant role of the mammalian ...
|
|
25454
|
Interactions of quinones with thioredoxin reductase: a challenge to the antioxidant role of the mammalian ...
|
|
25455
|
Interactions of quinones with thioredoxin reductase: a challenge to the antioxidant role of the mammalian ...
|
|
25456
|
Interactions of quinones with thioredoxin reductase: a challenge to the antioxidant role of the mammalian ...
|
|
25457
|
Interactions of quinones with thioredoxin reductase: a challenge to the antioxidant role of the mammalian ...
|
|
25458
|
Interactions of quinones with thioredoxin reductase: a challenge to the antioxidant role of the mammalian ...
|
|
25459
|
Interactions of quinones with thioredoxin reductase: a challenge to the antioxidant role of the mammalian ...
|
|
25460
|
Interactions of quinones with thioredoxin reductase: a challenge to the antioxidant role of the mammalian ...
|
|
25461
|
Interactions of quinones with thioredoxin reductase: a challenge to the antioxidant role of the mammalian ...
|
|
25462
|
Interactions of quinones with thioredoxin reductase: a challenge to the antioxidant role of the mammalian ...
|
|
25463
|
Interactions of quinones with thioredoxin reductase: a challenge to the antioxidant role of the mammalian ...
|
|
25464
|
Interactions of quinones with thioredoxin reductase: a challenge to the antioxidant role of the mammalian ...
|
|
25465
|
Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina ...
|
|
25466
|
Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina ...
|
|
25467
|
Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina ...
|
|
25468
|
Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina ...
|
|
25469
|
Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina ...
|
|
25470
|
Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina ...
|
|
25471
|
Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina ...
|
|
25472
|
Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina ...
|
|
25473
|
Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina ...
|
|
25474
|
Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina ...
|
|
25475
|
Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina ...
|
|
25476
|
Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina ...
|
|
25477
|
Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina ...
|
|
25478
|
Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina ...
|
|
25479
|
Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina ...
|
|
25480
|
Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina ...
|
|
25481
|
Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina ...
|
|
25482
|
Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina ...
|
|
25483
|
Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina ...
|
|
25484
|
Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina ...
|
|
25485
|
Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina ...
|
|
25486
|
Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina ...
|
|
25487
|
Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina ...
|
|
25488
|
Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina ...
|
|
25489
|
Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina ...
|
|
25490
|
Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina ...
|
|
25491
|
Site-directed Mutational Analysis of Active Site Residues in the Acetate Kinase from Methanosarcina ...
|
|
25492
|
Purification and characterization of acid phosphatase from yellow lupin (Lupinus luteus) seeds
|
|
25493
|
Purification and characterization of acid phosphatase from yellow lupin (Lupinus luteus) seeds
|
|
25494
|
Purification and characterization of acid phosphatase from yellow lupin (Lupinus luteus) seeds
|
|
25495
|
Purification and characterization of acid phosphatase from yellow lupin (Lupinus luteus) seeds
|
|
25496
|
Purification and characterization of acid phosphatase from yellow lupin (Lupinus luteus) seeds
|
|
25497
|
Purification and characterization of acid phosphatase from yellow lupin (Lupinus luteus) seeds
|
|
25498
|
Purification and characterization of acid phosphatase from yellow lupin (Lupinus luteus) seeds
|
|
25499
|
Purification and characterization of acid phosphatase from yellow lupin (Lupinus luteus) seeds
|
|
25500
|
Purification and characterization of acid phosphatase from yellow lupin (Lupinus luteus) seeds
|
|
25501
|
Purification and characterization of acid phosphatase from yellow lupin (Lupinus luteus) seeds
|
|
25502
|
Purification and characterization of acid phosphatase from yellow lupin (Lupinus luteus) seeds
|
|
25503
|
Purification and characterization of acid phosphatase from yellow lupin (Lupinus luteus) seeds
|
|
25504
|
Purification and characterization of acid phosphatase from yellow lupin (Lupinus luteus) seeds
|
|
25505
|
Purification and characterization of acid phosphatase from yellow lupin (Lupinus luteus) seeds
|
|
25506
|
Purification and characterization of acid phosphatase from yellow lupin (Lupinus luteus) seeds
|
|
25507
|
Purification and characterization of acid phosphatase from yellow lupin (Lupinus luteus) seeds
|
|
25508
|
Purification and characterization of acid phosphatase from yellow lupin (Lupinus luteus) seeds
|
|
25509
|
Purification and characterization of acid phosphatase from yellow lupin (Lupinus luteus) seeds
|
|
25510
|
Purification and characterization of acid phosphatase from yellow lupin (Lupinus luteus) seeds
|
|
25511
|
Purification and characterization of acid phosphatase from yellow lupin (Lupinus luteus) seeds
|
|
25512
|
Purification and characterization of acid phosphatase from yellow lupin (Lupinus luteus) seeds
|
|
25513
|
Purification and characterization of acid phosphatase from yellow lupin (Lupinus luteus) seeds
|
|
25514
|
Purification and characterization of acid phosphatase from yellow lupin (Lupinus luteus) seeds
|
|
25515
|
Differential and special properties of the major human UGT1-encoded gastrointestinal ...
|
|
25516
|
Differential and special properties of the major human UGT1-encoded gastrointestinal ...
|
|
25517
|
Differential and special properties of the major human UGT1-encoded gastrointestinal ...
|
|
25518
|
Differential and special properties of the major human UGT1-encoded gastrointestinal ...
|
|
25519
|
Differential and special properties of the major human UGT1-encoded gastrointestinal ...
|
|
25520
|
Differential and special properties of the major human UGT1-encoded gastrointestinal ...
|
|
25521
|
Differential and special properties of the major human UGT1-encoded gastrointestinal ...
|
|
25522
|
Differential and special properties of the major human UGT1-encoded gastrointestinal ...
|
|
25523
|
Differential and special properties of the major human UGT1-encoded gastrointestinal ...
|
|
25524
|
Differential and special properties of the major human UGT1-encoded gastrointestinal ...
|
|
25525
|
Differential and special properties of the major human UGT1-encoded gastrointestinal ...
|
|
25526
|
Differential and special properties of the major human UGT1-encoded gastrointestinal ...
|
|
25527
|
Differential and special properties of the major human UGT1-encoded gastrointestinal ...
|
|
25528
|
Differential and special properties of the major human UGT1-encoded gastrointestinal ...
|
|
25529
|
Differential and special properties of the major human UGT1-encoded gastrointestinal ...
|
|
25530
|
Differential and special properties of the major human UGT1-encoded gastrointestinal ...
|
|
25531
|
A mutant Escherichia coli tyrosyl-tRNA synthetase utilizes the unnatural amino acid azatyrosine more ...
|
|
25532
|
A mutant Escherichia coli tyrosyl-tRNA synthetase utilizes the unnatural amino acid azatyrosine more ...
|
|
25533
|
A mutant Escherichia coli tyrosyl-tRNA synthetase utilizes the unnatural amino acid azatyrosine more ...
|
|
25534
|
A mutant Escherichia coli tyrosyl-tRNA synthetase utilizes the unnatural amino acid azatyrosine more ...
|
|
25535
|
Purification and properties of 4-aminobutyrate 2-ketoglutarate aminotransferase from pig liver
|
|
25536
|
Purification and properties of 4-aminobutyrate 2-ketoglutarate aminotransferase from pig liver
|
|
25537
|
Purification and properties of 4-aminobutyrate 2-ketoglutarate aminotransferase from pig liver
|
|
25538
|
Hydrophobic Amino Acid Residues in the Acceptor Binding Site Are Main Determinants for Reaction Mechanism and ...
|
|
25539
|
Hydrophobic Amino Acid Residues in the Acceptor Binding Site Are Main Determinants for Reaction Mechanism and ...
|
|
25540
|
Hydrophobic Amino Acid Residues in the Acceptor Binding Site Are Main Determinants for Reaction Mechanism and ...
|
|
25541
|
Hydrophobic Amino Acid Residues in the Acceptor Binding Site Are Main Determinants for Reaction Mechanism and ...
|
|
25542
|
Hydrophobic Amino Acid Residues in the Acceptor Binding Site Are Main Determinants for Reaction Mechanism and ...
|
|
25543
|
Hydrophobic Amino Acid Residues in the Acceptor Binding Site Are Main Determinants for Reaction Mechanism and ...
|
|
25544
|
Hydrophobic Amino Acid Residues in the Acceptor Binding Site Are Main Determinants for Reaction Mechanism and ...
|
|
25545
|
Hydrophobic Amino Acid Residues in the Acceptor Binding Site Are Main Determinants for Reaction Mechanism and ...
|
|
25546
|
Hydrophobic Amino Acid Residues in the Acceptor Binding Site Are Main Determinants for Reaction Mechanism and ...
|
|
25547
|
Hydrophobic Amino Acid Residues in the Acceptor Binding Site Are Main Determinants for Reaction Mechanism and ...
|
|
25548
|
Hydrophobic Amino Acid Residues in the Acceptor Binding Site Are Main Determinants for Reaction Mechanism and ...
|
|
25549
|
Hydrophobic Amino Acid Residues in the Acceptor Binding Site Are Main Determinants for Reaction Mechanism and ...
|
|
25550
|
Hydrophobic Amino Acid Residues in the Acceptor Binding Site Are Main Determinants for Reaction Mechanism and ...
|
|
25551
|
Hydrophobic Amino Acid Residues in the Acceptor Binding Site Are Main Determinants for Reaction Mechanism and ...
|
|
25552
|
Hydrophobic Amino Acid Residues in the Acceptor Binding Site Are Main Determinants for Reaction Mechanism and ...
|
|
25553
|
Hydrophobic Amino Acid Residues in the Acceptor Binding Site Are Main Determinants for Reaction Mechanism and ...
|
|
25554
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25555
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25556
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25557
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25558
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25559
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25560
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25561
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25562
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25563
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25564
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25565
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25566
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25567
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25568
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25569
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25570
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25571
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25572
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25573
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25574
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25575
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25576
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25577
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25578
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25579
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25580
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25581
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25582
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25583
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25584
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25585
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25586
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25587
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25588
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25589
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25590
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25591
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25592
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25593
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25594
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25595
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25596
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25597
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25598
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25599
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25600
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25601
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25602
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25603
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25604
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25605
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25606
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25607
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25608
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25609
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25610
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25611
|
S-Adenosylmethionine Synthetase from Escherichia coli
|
|
25612
|
Sugar transport by the bacterial phosphotransferase system. Isolation and characterization of enzyme I from ...
|
|
25613
|
Sugar transport by the bacterial phosphotransferase system. Isolation and characterization of enzyme I from ...
|
|
25614
|
Sugar transport by the bacterial phosphotransferase system. Isolation and characterization of enzyme I from ...
|
|
25615
|
Sugar transport by the bacterial phosphotransferase system. Isolation and characterization of enzyme I from ...
|
|
25616
|
Sugar transport by the bacterial phosphotransferase system. Isolation and characterization of enzyme I from ...
|
|
25617
|
Sugar transport by the bacterial phosphotransferase system. Isolation and characterization of enzyme I from ...
|
|
25618
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25619
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25620
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25621
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25622
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25623
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25624
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25625
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25626
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25627
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25628
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25629
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25630
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25631
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25632
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25633
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25634
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25635
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25636
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25637
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25638
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25639
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25640
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25641
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25642
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25643
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25644
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25645
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25646
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25647
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25648
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25649
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25650
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25651
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25652
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25653
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25654
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25655
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25656
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25657
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25658
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25659
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25660
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25661
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25662
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25663
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25664
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25665
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25666
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25667
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25668
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25669
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25670
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25671
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25672
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25673
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25674
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25675
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25676
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25677
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25678
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25679
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25680
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25681
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25682
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25683
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25684
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25685
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25686
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25687
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25688
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25689
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25690
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25691
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25692
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25693
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25694
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25695
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25696
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25697
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25698
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25699
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25700
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25701
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25702
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25703
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25704
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25705
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25706
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25707
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25708
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25709
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25710
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25711
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25712
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25713
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25714
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25715
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25716
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25717
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25718
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25719
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25720
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25721
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25722
|
Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects
|
|
25723
|
Regulation of glucosylation of teichoic acid. II. Partial characterization of phosphoglucomutase in Bacillus ...
|
|
25724
|
Regulation of glucosylation of teichoic acid. II. Partial characterization of phosphoglucomutase in Bacillus ...
|
|
25725
|
Regulation of glucosylation of teichoic acid. II. Partial characterization of phosphoglucomutase in Bacillus ...
|
|
25726
|
Regulation of glucosylation of teichoic acid. II. Partial characterization of phosphoglucomutase in Bacillus ...
|
|
25727
|
Regulation of glucosylation of teichoic acid. II. Partial characterization of phosphoglucomutase in Bacillus ...
|
|
25728
|
Regulation of glucosylation of teichoic acid. II. Partial characterization of phosphoglucomutase in Bacillus ...
|
|
25729
|
Regulation of glucosylation of teichoic acid. II. Partial characterization of phosphoglucomutase in Bacillus ...
|
|
25730
|
Regulation of glucosylation of teichoic acid. II. Partial characterization of phosphoglucomutase in Bacillus ...
|
|
25731
|
Regulation of glucosylation of teichoic acid. II. Partial characterization of phosphoglucomutase in Bacillus ...
|
|
25732
|
Regulation of glucosylation of teichoic acid. II. Partial characterization of phosphoglucomutase in Bacillus ...
|
|
25733
|
Regulation of glucosylation of teichoic acid. II. Partial characterization of phosphoglucomutase in Bacillus ...
|
|
25734
|
Regulation of glucosylation of teichoic acid. II. Partial characterization of phosphoglucomutase in Bacillus ...
|
|
25735
|
Regulation of glucosylation of teichoic acid. II. Partial characterization of phosphoglucomutase in Bacillus ...
|
|
25736
|
Regulation of glucosylation of teichoic acid. II. Partial characterization of phosphoglucomutase in Bacillus ...
|
|
25737
|
Regulation of glucosylation of teichoic acid. II. Partial characterization of phosphoglucomutase in Bacillus ...
|
|
25738
|
Partial purification and characterization of sphingosine N-acyltransferase (ceramide synthase) from bovine ...
|
|
25739
|
Partial purification and characterization of sphingosine N-acyltransferase (ceramide synthase) from bovine ...
|
|
25740
|
Partial purification and characterization of sphingosine N-acyltransferase (ceramide synthase) from bovine ...
|
|
25741
|
Partial purification and characterization of sphingosine N-acyltransferase (ceramide synthase) from bovine ...
|
|
25742
|
Partial purification and characterization of sphingosine N-acyltransferase (ceramide synthase) from bovine ...
|
|
25743
|
Partial purification and characterization of sphingosine N-acyltransferase (ceramide synthase) from bovine ...
|
|
25744
|
Partial purification and characterization of sphingosine N-acyltransferase (ceramide synthase) from bovine ...
|
|
25745
|
Partial purification and characterization of sphingosine N-acyltransferase (ceramide synthase) from bovine ...
|
|
25746
|
Isolation and properties of the glycolytic enzymes from Zymomonas mobilis. The five enzymes from ...
|
|
25747
|
Isolation and properties of the glycolytic enzymes from Zymomonas mobilis. The five enzymes from ...
|
|
25748
|
Isolation and properties of the glycolytic enzymes from Zymomonas mobilis. The five enzymes from ...
|
|
25749
|
Isolation and properties of the glycolytic enzymes from Zymomonas mobilis. The five enzymes from ...
|
|
25750
|
Isolation and properties of the glycolytic enzymes from Zymomonas mobilis. The five enzymes from ...
|
|
25751
|
Isolation and properties of the glycolytic enzymes from Zymomonas mobilis. The five enzymes from ...
|
|
25752
|
Isolation and properties of the glycolytic enzymes from Zymomonas mobilis. The five enzymes from ...
|
|
25753
|
Human monocyte carboxylesterase. Purification and kinetics
|
|
25754
|
Human monocyte carboxylesterase. Purification and kinetics
|
|
25755
|
Human monocyte carboxylesterase. Purification and kinetics
|
|
25756
|
Human monocyte carboxylesterase. Purification and kinetics
|
|
25757
|
Human monocyte carboxylesterase. Purification and kinetics
|
|
25758
|
Human monocyte carboxylesterase. Purification and kinetics
|
|
25759
|
Human monocyte carboxylesterase. Purification and kinetics
|
|
25760
|
Purification and characterization of N-acetylglucosamine kinase from rat liver--comparison with ...
|
|
25761
|
Purification and characterization of N-acetylglucosamine kinase from rat liver--comparison with ...
|
|
25762
|
Enzyme characteristics of two distinct forms of mouse 3 beta-hydroxysteroid dehydrogenase/delta 5-delta ...
|
|
25763
|
Enzyme characteristics of two distinct forms of mouse 3 beta-hydroxysteroid dehydrogenase/delta 5-delta ...
|
|
25764
|
Enzyme characteristics of two distinct forms of mouse 3 beta-hydroxysteroid dehydrogenase/delta 5-delta ...
|
|
25765
|
Enzyme characteristics of two distinct forms of mouse 3 beta-hydroxysteroid dehydrogenase/delta 5-delta ...
|
|
25766
|
Enzyme characteristics of two distinct forms of mouse 3 beta-hydroxysteroid dehydrogenase/delta 5-delta ...
|
|
25767
|
Enzyme characteristics of two distinct forms of mouse 3 beta-hydroxysteroid dehydrogenase/delta 5-delta ...
|
|
25768
|
Enzyme characteristics of two distinct forms of mouse 3 beta-hydroxysteroid dehydrogenase/delta 5-delta ...
|
|
25769
|
Enzyme characteristics of two distinct forms of mouse 3 beta-hydroxysteroid dehydrogenase/delta 5-delta ...
|
|
25770
|
Thymidylate synthetase purified to homogeneity from human leukemic cells
|
|
25771
|
Purification and characterization of a rat brain aldehyde dehydrogenase able to metabolize ...
|
|
25772
|
Purification and characterization of a rat brain aldehyde dehydrogenase able to metabolize ...
|
|
25773
|
Purification and characterization of a rat brain aldehyde dehydrogenase able to metabolize ...
|
|
25774
|
Purification and characterization of a rat brain aldehyde dehydrogenase able to metabolize ...
|
|
25775
|
Purification and characterization of a rat brain aldehyde dehydrogenase able to metabolize ...
|
|
25776
|
Purification and characterization of a rat brain aldehyde dehydrogenase able to metabolize ...
|
|
25777
|
Cloning, expression and characterization of the Streptococcus pyogenes murE gene encoding a ...
|
|
25778
|
Cloning, expression and characterization of the Streptococcus pyogenes murE gene encoding a ...
|
|
25779
|
Cloning, expression and characterization of the Streptococcus pyogenes murE gene encoding a ...
|
|
25780
|
Site-directed mutagenesis of ATP binding residues of Biotin carboxylase
|
|
25781
|
Site-directed mutagenesis of ATP binding residues of Biotin carboxylase
|
|
25782
|
Site-directed mutagenesis of ATP binding residues of Biotin carboxylase
|
|
25783
|
Site-directed mutagenesis of ATP binding residues of Biotin carboxylase
|
|
25784
|
Site-directed mutagenesis of ATP binding residues of Biotin carboxylase
|
|
25785
|
Site-directed mutagenesis of ATP binding residues of Biotin carboxylase
|
|
25786
|
Site-directed mutagenesis of ATP binding residues of Biotin carboxylase
|
|
25787
|
Site-directed mutagenesis of ATP binding residues of Biotin carboxylase
|
|
25788
|
Site-directed mutagenesis of ATP binding residues of Biotin carboxylase
|
|
25789
|
Site-directed mutagenesis of ATP binding residues of Biotin carboxylase
|
|
25790
|
Site-directed mutagenesis of ATP binding residues of Biotin carboxylase
|
|
25791
|
Site-directed mutagenesis of ATP binding residues of Biotin carboxylase
|
|
25792
|
Site-directed mutagenesis of ATP binding residues of Biotin carboxylase
|
|
25793
|
Site-directed mutagenesis of ATP binding residues of Biotin carboxylase
|
|
25794
|
Site-directed mutagenesis of ATP binding residues of Biotin carboxylase
|
|
25795
|
Site-directed mutagenesis of ATP binding residues of Biotin carboxylase
|
|
25796
|
Site-directed mutagenesis of ATP binding residues of Biotin carboxylase
|
|
25797
|
Site-directed mutagenesis of ATP binding residues of Biotin carboxylase
|
|
25798
|
Site-directed mutagenesis of ATP binding residues of Biotin carboxylase
|
|
25799
|
Site-directed mutagenesis of ATP binding residues of Biotin carboxylase
|
|
25800
|
Site-directed mutagenesis of ATP binding residues of Biotin carboxylase
|
|
25801
|
Breaking symmetry: mutations engineered into R67 dihydrofolate reductase, a D2 symmetric homotetramer ...
|
|
25802
|
Breaking symmetry: mutations engineered into R67 dihydrofolate reductase, a D2 symmetric homotetramer ...
|
|
25803
|
Breaking symmetry: mutations engineered into R67 dihydrofolate reductase, a D2 symmetric homotetramer ...
|
|
25804
|
Breaking symmetry: mutations engineered into R67 dihydrofolate reductase, a D2 symmetric homotetramer ...
|
|
25805
|
Breaking symmetry: mutations engineered into R67 dihydrofolate reductase, a D2 symmetric homotetramer ...
|
|
25806
|
Breaking symmetry: mutations engineered into R67 dihydrofolate reductase, a D2 symmetric homotetramer ...
|
|
25807
|
Breaking symmetry: mutations engineered into R67 dihydrofolate reductase, a D2 symmetric homotetramer ...
|
|
25808
|
Breaking symmetry: mutations engineered into R67 dihydrofolate reductase, a D2 symmetric homotetramer ...
|
|
25809
|
Breaking symmetry: mutations engineered into R67 dihydrofolate reductase, a D2 symmetric homotetramer ...
|
|
25810
|
Breaking symmetry: mutations engineered into R67 dihydrofolate reductase, a D2 symmetric homotetramer ...
|
|
25811
|
Breaking symmetry: mutations engineered into R67 dihydrofolate reductase, a D2 symmetric homotetramer ...
|
|
25812
|
Breaking symmetry: mutations engineered into R67 dihydrofolate reductase, a D2 symmetric homotetramer ...
|
|
25813
|
Identification and functional characterization of an active-site lysine in mevalonate kinase
|
|
25814
|
Identification and functional characterization of an active-site lysine in mevalonate kinase
|
|
25815
|
Identification and functional characterization of an active-site lysine in mevalonate kinase
|
|
25816
|
Identification and functional characterization of an active-site lysine in mevalonate kinase
|
|
25817
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25818
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25819
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25820
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25821
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25822
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25823
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25824
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25825
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25826
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25827
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25828
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25829
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25830
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25831
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25832
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25833
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25834
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25835
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25836
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25837
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25838
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25839
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25840
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25841
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25842
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25843
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25844
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25845
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25846
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25847
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25848
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25849
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25850
|
Purification and characterization of a novel dimeric 20 alpha-hydroxysteroid dehydrogenase from Tetrahymena ...
|
|
25851
|
Solution structure, enzymatic, and non-enzymatic reactivity of 3-isoadenosylcobalamin, a structural isomer of ...
|
|
25852
|
Solution structure, enzymatic, and non-enzymatic reactivity of 3-isoadenosylcobalamin, a structural isomer of ...
|
|
25853
|
Solution structure, enzymatic, and non-enzymatic reactivity of 3-isoadenosylcobalamin, a structural isomer of ...
|
|
25854
|
Solution structure, enzymatic, and non-enzymatic reactivity of 3-isoadenosylcobalamin, a structural isomer of ...
|
|
25855
|
Solution structure, enzymatic, and non-enzymatic reactivity of 3-isoadenosylcobalamin, a structural isomer of ...
|
|
25856
|
Solution structure, enzymatic, and non-enzymatic reactivity of 3-isoadenosylcobalamin, a structural isomer of ...
|
|
25857
|
Solution structure, enzymatic, and non-enzymatic reactivity of 3-isoadenosylcobalamin, a structural isomer of ...
|
|
25858
|
Solution structure, enzymatic, and non-enzymatic reactivity of 3-isoadenosylcobalamin, a structural isomer of ...
|
|
25859
|
A phenylalanine codon deletion at the UGT1 gene complex locus of a Crigler-Najjar type I patient generates a ...
|
|
25860
|
A phenylalanine codon deletion at the UGT1 gene complex locus of a Crigler-Najjar type I patient generates a ...
|
|
25861
|
A phenylalanine codon deletion at the UGT1 gene complex locus of a Crigler-Najjar type I patient generates a ...
|
|
25862
|
Molecular cloning and characterization of scrB, the structural gene for the Streptococcus mutans ...
|
|
25863
|
Molecular cloning and characterization of scrB, the structural gene for the Streptococcus mutans ...
|
|
25864
|
Aspartokinase of Streptococcus mutans: purification, properties, and regulation
|
|
25865
|
Aspartokinase of Streptococcus mutans: purification, properties, and regulation
|
|
25866
|
Aspartokinase of Streptococcus mutans: purification, properties, and regulation
|
|
25867
|
Characterization of retinaldehyde dehydrogenase 3
|
|
25868
|
Characterization of retinaldehyde dehydrogenase 3
|
|
25869
|
Characterization of retinaldehyde dehydrogenase 3
|
|
25870
|
Characterization of retinaldehyde dehydrogenase 3
|
|
25871
|
Characterization of retinaldehyde dehydrogenase 3
|
|
25872
|
Characterization of retinaldehyde dehydrogenase 3
|
|
25873
|
Characterization of retinaldehyde dehydrogenase 3
|
|
25874
|
Characterization of retinaldehyde dehydrogenase 3
|
|
25875
|
Characterization of retinaldehyde dehydrogenase 3
|
|
25876
|
Characterization of retinaldehyde dehydrogenase 3
|
|
25877
|
Characterization of retinaldehyde dehydrogenase 3
|
|
25878
|
Characterization of retinaldehyde dehydrogenase 3
|
|
25879
|
Glutamine synthetase and glutamine synthetase-like protein from human brain: purification and comparative ...
|
|
25880
|
Glutamine synthetase and glutamine synthetase-like protein from human brain: purification and comparative ...
|
|
25881
|
Glutamine synthetase and glutamine synthetase-like protein from human brain: purification and comparative ...
|
|
25882
|
Glutamine synthetase and glutamine synthetase-like protein from human brain: purification and comparative ...
|
|
25883
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25884
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25885
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25886
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25887
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25888
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25889
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25890
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25891
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25892
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25893
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25894
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25895
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25896
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25897
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25898
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25899
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25900
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25901
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25902
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25903
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25904
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25905
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25906
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25907
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25908
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25909
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25910
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25911
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25912
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25913
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25914
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25915
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25916
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25917
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25918
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25919
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25920
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25921
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25922
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25923
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25924
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25925
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25926
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25927
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25928
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25929
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25930
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25931
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25932
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25933
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25934
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25935
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25936
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25937
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25938
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25939
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25940
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25941
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25942
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25943
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25944
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25945
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25946
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25947
|
The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity
|
|
25948
|
Studies on streptococcal NAD-glycohydrolase. Copurification of streptodornase A
|
|
25949
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25950
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25951
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25952
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25953
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25954
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25955
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25956
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25957
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25958
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25959
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25960
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25961
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25962
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25963
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25964
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25965
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25966
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25967
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25968
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25969
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25970
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25971
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25972
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25973
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25974
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25975
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25976
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25977
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25978
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25979
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25980
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25981
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25982
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25983
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25984
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25985
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25986
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25987
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25988
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25989
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25990
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25991
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25992
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25993
|
Nucleotide specificity of HIV-1 reverse transcriptases with amino acid substitutions affecting Ala-114
|
|
25994
|
Probing the catalytic mechanism of prephenate dehydratase by site-directed mutagenesis of the Escherichia coli ...
|
|
25995
|
Probing the catalytic mechanism of prephenate dehydratase by site-directed mutagenesis of the Escherichia coli ...
|
|
25996
|
Probing the catalytic mechanism of prephenate dehydratase by site-directed mutagenesis of the Escherichia coli ...
|
|
25997
|
Probing the catalytic mechanism of prephenate dehydratase by site-directed mutagenesis of the Escherichia coli ...
|
|
25998
|
Probing the catalytic mechanism of prephenate dehydratase by site-directed mutagenesis of the Escherichia coli ...
|
|
25999
|
Probing the catalytic mechanism of prephenate dehydratase by site-directed mutagenesis of the Escherichia coli ...
|
|
26000
|
Probing the catalytic mechanism of prephenate dehydratase by site-directed mutagenesis of the Escherichia coli ...
|





 If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.
If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.