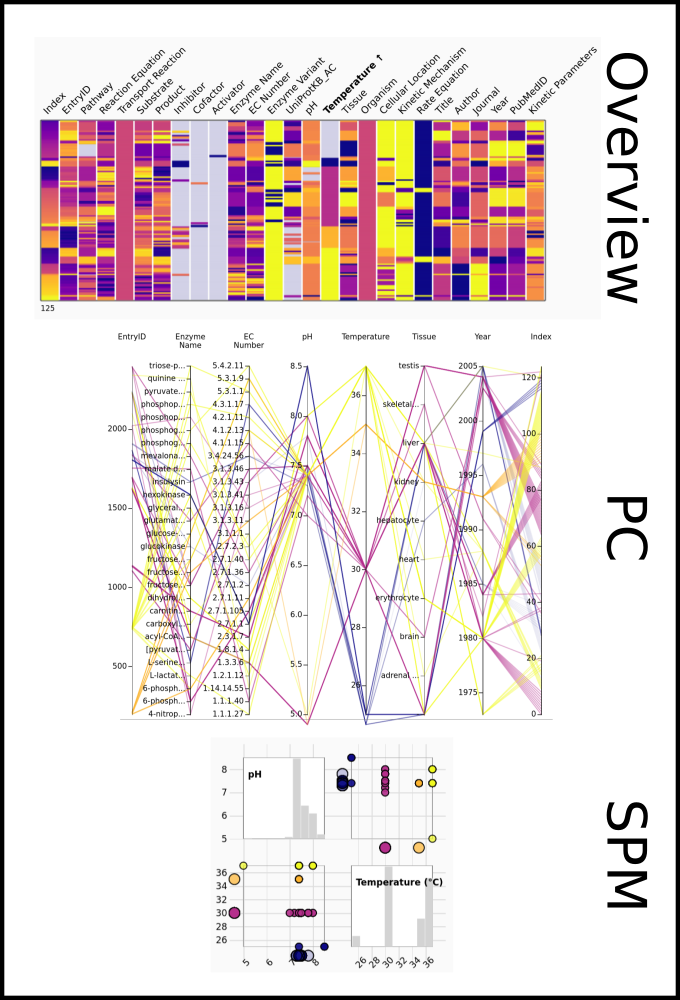
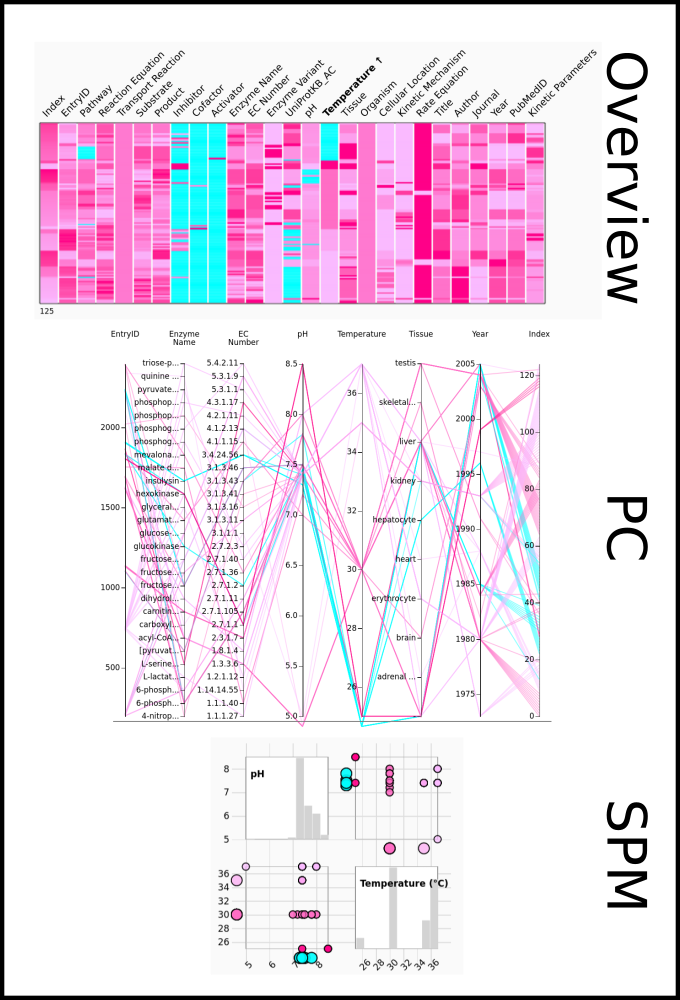
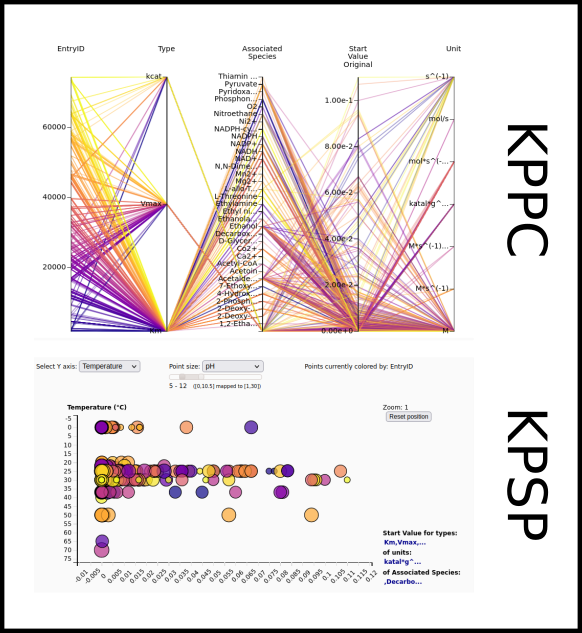
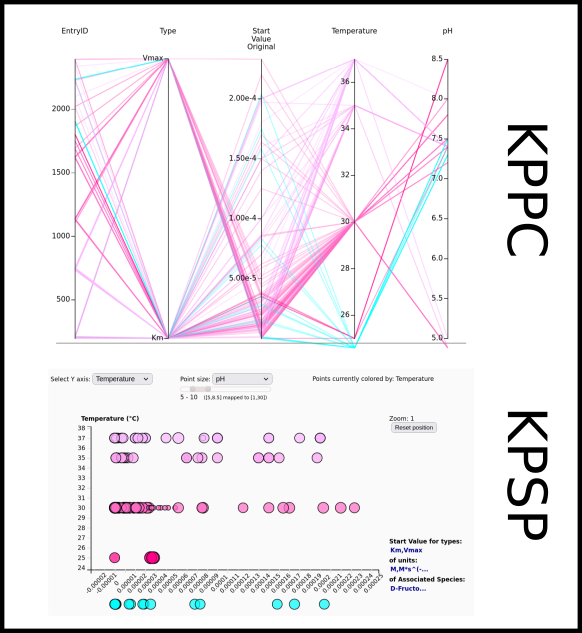
To improve SABIO-RK data interoperability, semantic markup was added to web pages as described and defined by
This structured information makes it easier to discover, collate and analyse our data.
|
24001
|
Metabolism in human cells of the D and L enantiomers of the carbocyclic analog of 2`-deoxyguanosine: substrate ...
|
|
24002
|
Metabolism in human cells of the D and L enantiomers of the carbocyclic analog of 2`-deoxyguanosine: substrate ...
|
|
24003
|
Metabolism in human cells of the D and L enantiomers of the carbocyclic analog of 2`-deoxyguanosine: substrate ...
|
|
24004
|
Metabolism in human cells of the D and L enantiomers of the carbocyclic analog of 2`-deoxyguanosine: substrate ...
|
|
24005
|
Metabolism in human cells of the D and L enantiomers of the carbocyclic analog of 2`-deoxyguanosine: substrate ...
|
|
24006
|
Metabolism in human cells of the D and L enantiomers of the carbocyclic analog of 2`-deoxyguanosine: substrate ...
|
|
24007
|
Synthesis and cleavage of octulose bisphosphates with liver and muscle aldolases
|
|
24008
|
Synthesis and cleavage of octulose bisphosphates with liver and muscle aldolases
|
|
24009
|
Synthesis and cleavage of octulose bisphosphates with liver and muscle aldolases
|
|
24010
|
Synthesis and cleavage of octulose bisphosphates with liver and muscle aldolases
|
|
24011
|
Synthesis and cleavage of octulose bisphosphates with liver and muscle aldolases
|
|
24012
|
Synthesis and cleavage of octulose bisphosphates with liver and muscle aldolases
|
|
24013
|
Synthesis and cleavage of octulose bisphosphates with liver and muscle aldolases
|
|
24014
|
Synthesis and cleavage of octulose bisphosphates with liver and muscle aldolases
|
|
24015
|
Synthesis and cleavage of octulose bisphosphates with liver and muscle aldolases
|
|
24016
|
Synthesis and cleavage of octulose bisphosphates with liver and muscle aldolases
|
|
24017
|
Transketolase A of Escherichia coli K12. Purification and properties of the enzyme from recombinant strains
|
|
24018
|
Transketolase A of Escherichia coli K12. Purification and properties of the enzyme from recombinant strains
|
|
24019
|
Transketolase A of Escherichia coli K12. Purification and properties of the enzyme from recombinant strains
|
|
24020
|
Transketolase A of Escherichia coli K12. Purification and properties of the enzyme from recombinant strains
|
|
24021
|
Transketolase A of Escherichia coli K12. Purification and properties of the enzyme from recombinant strains
|
|
24022
|
Transketolase A of Escherichia coli K12. Purification and properties of the enzyme from recombinant strains
|
|
24023
|
Transketolase A of Escherichia coli K12. Purification and properties of the enzyme from recombinant strains
|
|
24024
|
Transketolase A of Escherichia coli K12. Purification and properties of the enzyme from recombinant strains
|
|
24025
|
Transketolase A of Escherichia coli K12. Purification and properties of the enzyme from recombinant strains
|
|
24026
|
Transketolase A of Escherichia coli K12. Purification and properties of the enzyme from recombinant strains
|
|
24027
|
Transketolase A of Escherichia coli K12. Purification and properties of the enzyme from recombinant strains
|
|
24028
|
Transketolase A of Escherichia coli K12. Purification and properties of the enzyme from recombinant strains
|
|
24029
|
Transketolase A of Escherichia coli K12. Purification and properties of the enzyme from recombinant strains
|
|
24030
|
Transketolase A of Escherichia coli K12. Purification and properties of the enzyme from recombinant strains
|
|
24031
|
Transketolase A of Escherichia coli K12. Purification and properties of the enzyme from recombinant strains
|
|
24032
|
Transketolase A of Escherichia coli K12. Purification and properties of the enzyme from recombinant strains
|
|
24033
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24034
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24035
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24036
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24037
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24038
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24039
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24040
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24041
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24042
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24043
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24044
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24045
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24046
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24047
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24048
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24049
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24050
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24051
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24052
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24053
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24054
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24055
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24056
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24057
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24058
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24059
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24060
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24061
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24062
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24063
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24064
|
Identification of the histidine ligands to the binuclear metal center of phosphotriesterase by site-directed ...
|
|
24065
|
Kinetic isotope effects and ligand binding in PQQ-dependent methanol dehydrogenase
|
|
24066
|
Kinetic isotope effects and ligand binding in PQQ-dependent methanol dehydrogenase
|
|
24067
|
Kinetic isotope effects and ligand binding in PQQ-dependent methanol dehydrogenase
|
|
24068
|
Kinetic isotope effects and ligand binding in PQQ-dependent methanol dehydrogenase
|
|
24069
|
Kinetic isotope effects and ligand binding in PQQ-dependent methanol dehydrogenase
|
|
24070
|
Kinetic isotope effects and ligand binding in PQQ-dependent methanol dehydrogenase
|
|
24071
|
Kinetic isotope effects and ligand binding in PQQ-dependent methanol dehydrogenase
|
|
24072
|
Kinetic isotope effects and ligand binding in PQQ-dependent methanol dehydrogenase
|
|
24073
|
Kinetic isotope effects and ligand binding in PQQ-dependent methanol dehydrogenase
|
|
24074
|
Kinetic isotope effects and ligand binding in PQQ-dependent methanol dehydrogenase
|
|
24075
|
Kinetic isotope effects and ligand binding in PQQ-dependent methanol dehydrogenase
|
|
24076
|
Kinetic isotope effects and ligand binding in PQQ-dependent methanol dehydrogenase
|
|
24077
|
Kinetic isotope effects and ligand binding in PQQ-dependent methanol dehydrogenase
|
|
24078
|
Kinetic isotope effects and ligand binding in PQQ-dependent methanol dehydrogenase
|
|
24079
|
Kinetic isotope effects and ligand binding in PQQ-dependent methanol dehydrogenase
|
|
24080
|
Kinetic isotope effects and ligand binding in PQQ-dependent methanol dehydrogenase
|
|
24081
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24082
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24083
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24084
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24085
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24086
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24087
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24088
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24089
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24090
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24091
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24092
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24093
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24094
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24095
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24096
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24097
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24098
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24099
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24100
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24101
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24102
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24103
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24104
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24105
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24106
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24107
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24108
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24109
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24110
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24111
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24112
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24113
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24114
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24115
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24116
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24117
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24118
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24119
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24120
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24121
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24122
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24123
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24124
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24125
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24126
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24127
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24128
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24129
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24130
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24131
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24132
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24133
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24134
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24135
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24136
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24137
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24138
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24139
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24140
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24141
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24142
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24143
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24144
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24145
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24146
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24147
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24148
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24149
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24150
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24151
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24152
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24153
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24154
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24155
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24156
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24157
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24158
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24159
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24160
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24161
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24162
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24163
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24164
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24165
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24166
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24167
|
Activity profiles of deoxynucleoside kinases and 5`-nucleotidases in cultured adipocytes and myoblastic cells: ...
|
|
24168
|
Improved Catalytic Properties of Halohydrin Dehalogenase by Modification of the Halide-Binding Site
|
|
24169
|
Improved Catalytic Properties of Halohydrin Dehalogenase by Modification of the Halide-Binding Site
|
|
24170
|
Improved Catalytic Properties of Halohydrin Dehalogenase by Modification of the Halide-Binding Site
|
|
24171
|
Improved Catalytic Properties of Halohydrin Dehalogenase by Modification of the Halide-Binding Site
|
|
24172
|
Improved Catalytic Properties of Halohydrin Dehalogenase by Modification of the Halide-Binding Site
|
|
24173
|
Improved Catalytic Properties of Halohydrin Dehalogenase by Modification of the Halide-Binding Site
|
|
24174
|
Improved Catalytic Properties of Halohydrin Dehalogenase by Modification of the Halide-Binding Site
|
|
24175
|
Improved Catalytic Properties of Halohydrin Dehalogenase by Modification of the Halide-Binding Site
|
|
24176
|
Improved Catalytic Properties of Halohydrin Dehalogenase by Modification of the Halide-Binding Site
|
|
24177
|
Improved Catalytic Properties of Halohydrin Dehalogenase by Modification of the Halide-Binding Site
|
|
24178
|
Improved Catalytic Properties of Halohydrin Dehalogenase by Modification of the Halide-Binding Site
|
|
24179
|
Improved Catalytic Properties of Halohydrin Dehalogenase by Modification of the Halide-Binding Site
|
|
24180
|
Improved Catalytic Properties of Halohydrin Dehalogenase by Modification of the Halide-Binding Site
|
|
24181
|
Improved Catalytic Properties of Halohydrin Dehalogenase by Modification of the Halide-Binding Site
|
|
24182
|
Improved Catalytic Properties of Halohydrin Dehalogenase by Modification of the Halide-Binding Site
|
|
24183
|
Structural studies on a mitochondrial glyoxalase II
|
|
24184
|
Kinetic mechanism and enantioselectivity of halohydrin dehalogenase from Agrobacterium radiobacter
|
|
24185
|
Kinetic mechanism and enantioselectivity of halohydrin dehalogenase from Agrobacterium radiobacter
|
|
24186
|
Kinetic mechanism and enantioselectivity of halohydrin dehalogenase from Agrobacterium radiobacter
|
|
24187
|
Kinetic mechanism and enantioselectivity of halohydrin dehalogenase from Agrobacterium radiobacter
|
|
24188
|
Kinetic mechanism and enantioselectivity of halohydrin dehalogenase from Agrobacterium radiobacter
|
|
24189
|
Kinetic mechanism and enantioselectivity of halohydrin dehalogenase from Agrobacterium radiobacter
|
|
24190
|
Kinetic mechanism and enantioselectivity of halohydrin dehalogenase from Agrobacterium radiobacter
|
|
24191
|
Kinetic mechanism and enantioselectivity of halohydrin dehalogenase from Agrobacterium radiobacter
|
|
24192
|
Kinetic mechanism and enantioselectivity of halohydrin dehalogenase from Agrobacterium radiobacter
|
|
24193
|
Kinetic mechanism and enantioselectivity of halohydrin dehalogenase from Agrobacterium radiobacter
|
|
24194
|
Kinetic mechanism and enantioselectivity of halohydrin dehalogenase from Agrobacterium radiobacter
|
|
24195
|
Kinetic mechanism and enantioselectivity of halohydrin dehalogenase from Agrobacterium radiobacter
|
|
24196
|
Kinetic mechanism and enantioselectivity of halohydrin dehalogenase from Agrobacterium radiobacter
|
|
24197
|
Kinetic mechanism and enantioselectivity of halohydrin dehalogenase from Agrobacterium radiobacter
|
|
24198
|
Kinetic mechanism and enantioselectivity of halohydrin dehalogenase from Agrobacterium radiobacter
|
|
24199
|
Characterization of the gene encoding glyoxalase II from Leishmania donovani: a potential target for ...
|
|
24200
|
Stimulus-secretion coupling of arginine-induced insulin release: metabolism of L-arginine and L-omithine in ...
|
|
24201
|
Mechanism of rabbit muscle enolase: identification of the rate-limiting steps and the site of Li+ inhibition
|
|
24202
|
Mechanism of rabbit muscle enolase: identification of the rate-limiting steps and the site of Li+ inhibition
|
|
24203
|
Mechanism of rabbit muscle enolase: identification of the rate-limiting steps and the site of Li+ inhibition
|
|
24204
|
Mechanism of rabbit muscle enolase: identification of the rate-limiting steps and the site of Li+ inhibition
|
|
24205
|
Mechanism of rabbit muscle enolase: identification of the rate-limiting steps and the site of Li+ inhibition
|
|
24206
|
Mechanism of rabbit muscle enolase: identification of the rate-limiting steps and the site of Li+ inhibition
|
|
24207
|
Chemical rescue by guanidine derivatives of an arginine-substituted site-directed mutant of Escherichia coli ...
|
|
24208
|
Chemical rescue by guanidine derivatives of an arginine-substituted site-directed mutant of Escherichia coli ...
|
|
24209
|
Chemical rescue by guanidine derivatives of an arginine-substituted site-directed mutant of Escherichia coli ...
|
|
24210
|
Chemical rescue by guanidine derivatives of an arginine-substituted site-directed mutant of Escherichia coli ...
|
|
24211
|
Chemical rescue by guanidine derivatives of an arginine-substituted site-directed mutant of Escherichia coli ...
|
|
24212
|
Chemical rescue by guanidine derivatives of an arginine-substituted site-directed mutant of Escherichia coli ...
|
|
24213
|
Chemical rescue by guanidine derivatives of an arginine-substituted site-directed mutant of Escherichia coli ...
|
|
24214
|
Chemical rescue by guanidine derivatives of an arginine-substituted site-directed mutant of Escherichia coli ...
|
|
24215
|
Chemical rescue by guanidine derivatives of an arginine-substituted site-directed mutant of Escherichia coli ...
|
|
24216
|
Chemical rescue by guanidine derivatives of an arginine-substituted site-directed mutant of Escherichia coli ...
|
|
24217
|
Chemical rescue by guanidine derivatives of an arginine-substituted site-directed mutant of Escherichia coli ...
|
|
24218
|
Chemical rescue by guanidine derivatives of an arginine-substituted site-directed mutant of Escherichia coli ...
|
|
24219
|
Chemical rescue by guanidine derivatives of an arginine-substituted site-directed mutant of Escherichia coli ...
|
|
24220
|
Chemical rescue by guanidine derivatives of an arginine-substituted site-directed mutant of Escherichia coli ...
|
|
24221
|
Chemical rescue by guanidine derivatives of an arginine-substituted site-directed mutant of Escherichia coli ...
|
|
24222
|
Chemical rescue by guanidine derivatives of an arginine-substituted site-directed mutant of Escherichia coli ...
|
|
24223
|
Chemical rescue by guanidine derivatives of an arginine-substituted site-directed mutant of Escherichia coli ...
|
|
24224
|
Chemical rescue by guanidine derivatives of an arginine-substituted site-directed mutant of Escherichia coli ...
|
|
24225
|
Chemical rescue by guanidine derivatives of an arginine-substituted site-directed mutant of Escherichia coli ...
|
|
24226
|
Chemical rescue by guanidine derivatives of an arginine-substituted site-directed mutant of Escherichia coli ...
|
|
24227
|
Chemical rescue by guanidine derivatives of an arginine-substituted site-directed mutant of Escherichia coli ...
|
|
24228
|
Chemical rescue by guanidine derivatives of an arginine-substituted site-directed mutant of Escherichia coli ...
|
|
24229
|
Chemical rescue by guanidine derivatives of an arginine-substituted site-directed mutant of Escherichia coli ...
|
|
24230
|
Characterization of the structure and properties of the His 62-->Ala and Arg 38-->Ala mutants of yeast ...
|
|
24231
|
Characterization of the structure and properties of the His 62-->Ala and Arg 38-->Ala mutants of yeast ...
|
|
24232
|
Characterization of the structure and properties of the His 62-->Ala and Arg 38-->Ala mutants of yeast ...
|
|
24233
|
Characterization of the structure and properties of the His 62-->Ala and Arg 38-->Ala mutants of yeast ...
|
|
24234
|
Characterization of the structure and properties of the His 62-->Ala and Arg 38-->Ala mutants of yeast ...
|
|
24235
|
Characterization of the structure and properties of the His 62-->Ala and Arg 38-->Ala mutants of yeast ...
|
|
24236
|
Characterization of the structure and properties of the His 62-->Ala and Arg 38-->Ala mutants of yeast ...
|
|
24237
|
Characterization of the structure and properties of the His 62-->Ala and Arg 38-->Ala mutants of yeast ...
|
|
24238
|
Characterization of the structure and properties of the His 62-->Ala and Arg 38-->Ala mutants of yeast ...
|
|
24239
|
Characterization of the structure and properties of the His 62-->Ala and Arg 38-->Ala mutants of yeast ...
|
|
24240
|
Characterization of the structure and properties of the His 62-->Ala and Arg 38-->Ala mutants of yeast ...
|
|
24241
|
Functional studies of yeast-expressed human heart muscle carnitine palmitoyltransferase I
|
|
24242
|
Functional studies of yeast-expressed human heart muscle carnitine palmitoyltransferase I
|
|
24243
|
Functional studies of yeast-expressed human heart muscle carnitine palmitoyltransferase I
|
|
24244
|
Insights into the mechanism of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase (Phe) from Escherichia ...
|
|
24245
|
Insights into the mechanism of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase (Phe) from Escherichia ...
|
|
24246
|
Insights into the mechanism of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase (Phe) from Escherichia ...
|
|
24247
|
Insights into the mechanism of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase (Phe) from Escherichia ...
|
|
24248
|
Insights into the mechanism of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase (Phe) from Escherichia ...
|
|
24249
|
Insights into the mechanism of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase (Phe) from Escherichia ...
|
|
24250
|
Insights into the mechanism of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase (Phe) from Escherichia ...
|
|
24251
|
Insights into the mechanism of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase (Phe) from Escherichia ...
|
|
24252
|
Molecular cloning, expression, and characterization of myo-inositol oxygenase from mouse, rat, and human ...
|
|
24253
|
Molecular cloning, expression, and characterization of myo-inositol oxygenase from mouse, rat, and human ...
|
|
24254
|
Molecular cloning, expression, and characterization of myo-inositol oxygenase from mouse, rat, and human ...
|
|
24255
|
Molecular cloning, expression, and characterization of myo-inositol oxygenase from mouse, rat, and human ...
|
|
24256
|
Molecular cloning, expression, and characterization of myo-inositol oxygenase from mouse, rat, and human ...
|
|
24257
|
Molecular cloning, expression, and characterization of myo-inositol oxygenase from mouse, rat, and human ...
|
|
24258
|
myo-Inositol oxygenase: molecular cloning and expression of a unique enzyme that oxidizes myo-inositol and ...
|
|
24259
|
myo-Inositol oxygenase: molecular cloning and expression of a unique enzyme that oxidizes myo-inositol and ...
|
|
24260
|
Differences in the betaC-S lyase activities of viridans group streptococci
|
|
24261
|
Differences in the betaC-S lyase activities of viridans group streptococci
|
|
24262
|
Differences in the betaC-S lyase activities of viridans group streptococci
|
|
24263
|
Differences in the betaC-S lyase activities of viridans group streptococci
|
|
24264
|
Differences in the betaC-S lyase activities of viridans group streptococci
|
|
24265
|
Differences in the betaC-S lyase activities of viridans group streptococci
|
|
24266
|
Differences in the betaC-S lyase activities of viridans group streptococci
|
|
24267
|
Differences in the betaC-S lyase activities of viridans group streptococci
|
|
24268
|
Differences in the betaC-S lyase activities of viridans group streptococci
|
|
24269
|
Differences in the betaC-S lyase activities of viridans group streptococci
|
|
24270
|
Differences in the betaC-S lyase activities of viridans group streptococci
|
|
24271
|
Differences in the betaC-S lyase activities of viridans group streptococci
|
|
24272
|
Modelling of substrate binding to 3-phosphoglycerate kinase with analogues of 3-phosphoglycerate
|
|
24273
|
Modelling of substrate binding to 3-phosphoglycerate kinase with analogues of 3-phosphoglycerate
|
|
24274
|
Modelling of substrate binding to 3-phosphoglycerate kinase with analogues of 3-phosphoglycerate
|
|
24275
|
Modelling of substrate binding to 3-phosphoglycerate kinase with analogues of 3-phosphoglycerate
|
|
24276
|
Modelling of substrate binding to 3-phosphoglycerate kinase with analogues of 3-phosphoglycerate
|
|
24277
|
Cloning, nucleotide sequences, and enzymatic properties of glucose dehydrogenase isozymes from Bacillus ...
|
|
24278
|
Cloning, nucleotide sequences, and enzymatic properties of glucose dehydrogenase isozymes from Bacillus ...
|
|
24279
|
Cloning, nucleotide sequences, and enzymatic properties of glucose dehydrogenase isozymes from Bacillus ...
|
|
24280
|
Cloning, nucleotide sequences, and enzymatic properties of glucose dehydrogenase isozymes from Bacillus ...
|
|
24281
|
Cloning, nucleotide sequences, and enzymatic properties of glucose dehydrogenase isozymes from Bacillus ...
|
|
24282
|
Cloning, nucleotide sequences, and enzymatic properties of glucose dehydrogenase isozymes from Bacillus ...
|
|
24283
|
Cloning, nucleotide sequences, and enzymatic properties of glucose dehydrogenase isozymes from Bacillus ...
|
|
24284
|
Cloning, nucleotide sequences, and enzymatic properties of glucose dehydrogenase isozymes from Bacillus ...
|
|
24285
|
Cloning, nucleotide sequences, and enzymatic properties of glucose dehydrogenase isozymes from Bacillus ...
|
|
24286
|
Specific interaction of cytosolic and mitochondrial glyoxalase II with acidic phospholipids in form of ...
|
|
24287
|
Specific interaction of cytosolic and mitochondrial glyoxalase II with acidic phospholipids in form of ...
|
|
24288
|
Specific interaction of cytosolic and mitochondrial glyoxalase II with acidic phospholipids in form of ...
|
|
24289
|
Specific interaction of cytosolic and mitochondrial glyoxalase II with acidic phospholipids in form of ...
|
|
24290
|
Specific interaction of cytosolic and mitochondrial glyoxalase II with acidic phospholipids in form of ...
|
|
24291
|
Specific interaction of cytosolic and mitochondrial glyoxalase II with acidic phospholipids in form of ...
|
|
24292
|
Specific interaction of cytosolic and mitochondrial glyoxalase II with acidic phospholipids in form of ...
|
|
24293
|
Specific interaction of cytosolic and mitochondrial glyoxalase II with acidic phospholipids in form of ...
|
|
24294
|
Specific interaction of cytosolic and mitochondrial glyoxalase II with acidic phospholipids in form of ...
|
|
24295
|
Specific interaction of cytosolic and mitochondrial glyoxalase II with acidic phospholipids in form of ...
|
|
24296
|
The phosphate group of 3-phosphoglycerate accounts for conformational changes occurring on binding to ...
|
|
24297
|
The phosphate group of 3-phosphoglycerate accounts for conformational changes occurring on binding to ...
|
|
24298
|
The phosphate group of 3-phosphoglycerate accounts for conformational changes occurring on binding to ...
|
|
24299
|
The phosphate group of 3-phosphoglycerate accounts for conformational changes occurring on binding to ...
|
|
24300
|
The phosphate group of 3-phosphoglycerate accounts for conformational changes occurring on binding to ...
|
|
24301
|
The phosphate group of 3-phosphoglycerate accounts for conformational changes occurring on binding to ...
|
|
24302
|
The phosphate group of 3-phosphoglycerate accounts for conformational changes occurring on binding to ...
|
|
24303
|
The phosphate group of 3-phosphoglycerate accounts for conformational changes occurring on binding to ...
|
|
24304
|
The phosphate group of 3-phosphoglycerate accounts for conformational changes occurring on binding to ...
|
|
24305
|
Glyoxalase II of African trypanosomes is trypanothione-dependent
|
|
24306
|
Glyoxalase II of African trypanosomes is trypanothione-dependent
|
|
24307
|
Glyoxalase II of African trypanosomes is trypanothione-dependent
|
|
24308
|
Glyoxalase II of African trypanosomes is trypanothione-dependent
|
|
24309
|
Kinetic studies on the reaction catalyzed by phosphoglycerate kinase. II. The kinetic relationships between ...
|
|
24310
|
Kinetic studies on the reaction catalyzed by phosphoglycerate kinase. II. The kinetic relationships between ...
|
|
24311
|
Kinetic studies on the reaction catalyzed by phosphoglycerate kinase. II. The kinetic relationships between ...
|
|
24312
|
Kinetic studies on the reaction catalyzed by phosphoglycerate kinase. II. The kinetic relationships between ...
|
|
24313
|
Kinetic studies on the reaction catalyzed by phosphoglycerate kinase. II. The kinetic relationships between ...
|
|
24314
|
Kinetic studies on the reaction catalyzed by phosphoglycerate kinase. II. The kinetic relationships between ...
|
|
24315
|
Kinetic studies on the reaction catalyzed by phosphoglycerate kinase. II. The kinetic relationships between ...
|
|
24316
|
Kinetic studies on the reaction catalyzed by phosphoglycerate kinase. II. The kinetic relationships between ...
|
|
24317
|
Measurement of arginine transport in human erythrocytes using their intrinsic arginase activity: implications ...
|
|
24318
|
Measurement of arginine transport in human erythrocytes using their intrinsic arginase activity: implications ...
|
|
24319
|
Measurement of arginine transport in human erythrocytes using their intrinsic arginase activity: implications ...
|
|
24320
|
Measurement of arginine transport in human erythrocytes using their intrinsic arginase activity: implications ...
|
|
24321
|
Measurement of arginine transport in human erythrocytes using their intrinsic arginase activity: implications ...
|
|
24322
|
Measurement of arginine transport in human erythrocytes using their intrinsic arginase activity: implications ...
|
|
24323
|
Measurement of arginine transport in human erythrocytes using their intrinsic arginase activity: implications ...
|
|
24324
|
Measurement of arginine transport in human erythrocytes using their intrinsic arginase activity: implications ...
|
|
24325
|
The active-site residue tyr-175 in human glyoxalase II contributes to binding of glutathione derivatives
|
|
24326
|
The active-site residue tyr-175 in human glyoxalase II contributes to binding of glutathione derivatives
|
|
24327
|
Purification and characterization of the bacterial MraY translocase catalyzing the first membrane step of ...
|
|
24328
|
Purification and characterization of the bacterial MraY translocase catalyzing the first membrane step of ...
|
|
24329
|
Purification and characterization of the bacterial MraY translocase catalyzing the first membrane step of ...
|
|
24330
|
Characterization of human tissue carnosinase
|
|
24331
|
Characterization of human tissue carnosinase
|
|
24332
|
Pre-steady-state kinetics of Bacillus licheniformis 1,3-1,4-beta-glucanase: evidence for a regulatory binding ...
|
|
24333
|
Pre-steady-state kinetics of Bacillus licheniformis 1,3-1,4-beta-glucanase: evidence for a regulatory binding ...
|
|
24334
|
Pre-steady-state kinetics of Bacillus licheniformis 1,3-1,4-beta-glucanase: evidence for a regulatory binding ...
|
|
24335
|
Pre-steady-state kinetics of Bacillus licheniformis 1,3-1,4-beta-glucanase: evidence for a regulatory binding ...
|
|
24336
|
Pre-steady-state kinetics of Bacillus licheniformis 1,3-1,4-beta-glucanase: evidence for a regulatory binding ...
|
|
24337
|
Pre-steady-state kinetics of Bacillus licheniformis 1,3-1,4-beta-glucanase: evidence for a regulatory binding ...
|
|
24338
|
Pre-steady-state kinetics of Bacillus licheniformis 1,3-1,4-beta-glucanase: evidence for a regulatory binding ...
|
|
24339
|
Pre-steady-state kinetics of Bacillus licheniformis 1,3-1,4-beta-glucanase: evidence for a regulatory binding ...
|
|
24340
|
Pre-steady-state kinetics of Bacillus licheniformis 1,3-1,4-beta-glucanase: evidence for a regulatory binding ...
|
|
24341
|
Pre-steady-state kinetics of Bacillus licheniformis 1,3-1,4-beta-glucanase: evidence for a regulatory binding ...
|
|
24342
|
Pre-steady-state kinetics of Bacillus licheniformis 1,3-1,4-beta-glucanase: evidence for a regulatory binding ...
|
|
24343
|
Pre-steady-state kinetics of Bacillus licheniformis 1,3-1,4-beta-glucanase: evidence for a regulatory binding ...
|
|
24344
|
Pre-steady-state kinetics of Bacillus licheniformis 1,3-1,4-beta-glucanase: evidence for a regulatory binding ...
|
|
24345
|
Pre-steady-state kinetics of Bacillus licheniformis 1,3-1,4-beta-glucanase: evidence for a regulatory binding ...
|
|
24346
|
Pre-steady-state kinetics of Bacillus licheniformis 1,3-1,4-beta-glucanase: evidence for a regulatory binding ...
|
|
24347
|
Pre-steady-state kinetics of Bacillus licheniformis 1,3-1,4-beta-glucanase: evidence for a regulatory binding ...
|
|
24348
|
Pre-steady-state kinetics of Bacillus licheniformis 1,3-1,4-beta-glucanase: evidence for a regulatory binding ...
|
|
24349
|
Pre-steady-state kinetics of Bacillus licheniformis 1,3-1,4-beta-glucanase: evidence for a regulatory binding ...
|
|
24350
|
Pre-steady-state kinetics of Bacillus licheniformis 1,3-1,4-beta-glucanase: evidence for a regulatory binding ...
|
|
24351
|
Pre-steady-state kinetics of Bacillus licheniformis 1,3-1,4-beta-glucanase: evidence for a regulatory binding ...
|
|
24352
|
Presence of a plant-like glyoxalase II in Candida albicans
|
|
24353
|
Presence of a plant-like glyoxalase II in Candida albicans
|
|
24354
|
Presence of a plant-like glyoxalase II in Candida albicans
|
|
24355
|
Presence of a plant-like glyoxalase II in Candida albicans
|
|
24356
|
Presence of a plant-like glyoxalase II in Candida albicans
|
|
24357
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24358
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24359
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24360
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24361
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24362
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24363
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24364
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24365
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24366
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24367
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24368
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24369
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24370
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24371
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24372
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24373
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24374
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24375
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24376
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24377
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24378
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24379
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24380
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24381
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24382
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24383
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24384
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24385
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24386
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24387
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24388
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24389
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24390
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24391
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24392
|
Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 ...
|
|
24393
|
Effects of S-petasin on cyclic AMP production and enzyme activity of P450scc in rat zona ...
|
|
24394
|
Effects of S-petasin on cyclic AMP production and enzyme activity of P450scc in rat zona ...
|
|
24395
|
Effects of S-petasin on cyclic AMP production and enzyme activity of P450scc in rat zona ...
|
|
24396
|
Effects of S-petasin on cyclic AMP production and enzyme activity of P450scc in rat zona ...
|
|
24397
|
Characterization and Nucleotide Binding Properties of a Mutant Dihydropteridine Reductase Containing an ...
|
|
24398
|
Characterization and Nucleotide Binding Properties of a Mutant Dihydropteridine Reductase Containing an ...
|
|
24399
|
Characterization and Nucleotide Binding Properties of a Mutant Dihydropteridine Reductase Containing an ...
|
|
24400
|
Characterization and Nucleotide Binding Properties of a Mutant Dihydropteridine Reductase Containing an ...
|
|
24401
|
Characterization and Nucleotide Binding Properties of a Mutant Dihydropteridine Reductase Containing an ...
|
|
24402
|
Characterization and Nucleotide Binding Properties of a Mutant Dihydropteridine Reductase Containing an ...
|
|
24403
|
Characterization and Nucleotide Binding Properties of a Mutant Dihydropteridine Reductase Containing an ...
|
|
24404
|
Characterization and Nucleotide Binding Properties of a Mutant Dihydropteridine Reductase Containing an ...
|
|
24405
|
Formation and conversion of oxygen metabolites by Lactococcus lactis subsp. lactis ATCC 19435 under different ...
|
|
24406
|
The gene cluster for agmatine catabolism of Enterococcus faecalis: study of recombinant putrescine ...
|
|
24407
|
The gene cluster for agmatine catabolism of Enterococcus faecalis: study of recombinant putrescine ...
|
|
24408
|
The gene cluster for agmatine catabolism of Enterococcus faecalis: study of recombinant putrescine ...
|
|
24409
|
The gene cluster for agmatine catabolism of Enterococcus faecalis: study of recombinant putrescine ...
|
|
24410
|
Transforming growth factor-beta stimulates arginase activity in macrophages. Implications for the regulation ...
|
|
24411
|
Transforming growth factor-beta stimulates arginase activity in macrophages. Implications for the regulation ...
|
|
24412
|
In rat alveolar macrophages lipopolysaccharides exert divergent effects on the transport of the cationic amino ...
|
|
24413
|
In rat alveolar macrophages lipopolysaccharides exert divergent effects on the transport of the cationic amino ...
|
|
24414
|
In rat alveolar macrophages lipopolysaccharides exert divergent effects on the transport of the cationic amino ...
|
|
24415
|
In rat alveolar macrophages lipopolysaccharides exert divergent effects on the transport of the cationic amino ...
|
|
24416
|
In rat alveolar macrophages lipopolysaccharides exert divergent effects on the transport of the cationic amino ...
|
|
24417
|
In rat alveolar macrophages lipopolysaccharides exert divergent effects on the transport of the cationic amino ...
|
|
24418
|
In rat alveolar macrophages lipopolysaccharides exert divergent effects on the transport of the cationic amino ...
|
|
24419
|
In rat alveolar macrophages lipopolysaccharides exert divergent effects on the transport of the cationic amino ...
|
|
24420
|
In rat alveolar macrophages lipopolysaccharides exert divergent effects on the transport of the cationic amino ...
|
|
24421
|
In rat alveolar macrophages lipopolysaccharides exert divergent effects on the transport of the cationic amino ...
|
|
24422
|
In rat alveolar macrophages lipopolysaccharides exert divergent effects on the transport of the cationic amino ...
|
|
24423
|
In rat alveolar macrophages lipopolysaccharides exert divergent effects on the transport of the cationic amino ...
|
|
24424
|
In rat alveolar macrophages lipopolysaccharides exert divergent effects on the transport of the cationic amino ...
|
|
24425
|
Enzymatic reduction of alkyl and acyl derivatives of dihydroxyacetone phosphate by reduced pyridine ...
|
|
24426
|
Enzymatic reduction of alkyl and acyl derivatives of dihydroxyacetone phosphate by reduced pyridine ...
|
|
24427
|
Enzymatic reduction of alkyl and acyl derivatives of dihydroxyacetone phosphate by reduced pyridine ...
|
|
24428
|
Enzymatic reduction of alkyl and acyl derivatives of dihydroxyacetone phosphate by reduced pyridine ...
|
|
24429
|
Enzymatic reduction of alkyl and acyl derivatives of dihydroxyacetone phosphate by reduced pyridine ...
|
|
24430
|
Enzymatic reduction of alkyl and acyl derivatives of dihydroxyacetone phosphate by reduced pyridine ...
|
|
24431
|
Lactate dehydrogenase from Streptococcus mutans: purification, characterization, and crossed antigenicity with ...
|
|
24432
|
Lactate dehydrogenase from Streptococcus mutans: purification, characterization, and crossed antigenicity with ...
|
|
24433
|
Lactate dehydrogenase from Streptococcus mutans: purification, characterization, and crossed antigenicity with ...
|
|
24434
|
Lactate dehydrogenase from Streptococcus mutans: purification, characterization, and crossed antigenicity with ...
|
|
24435
|
Lactate dehydrogenase from Streptococcus mutans: purification, characterization, and crossed antigenicity with ...
|
|
24436
|
Lactate dehydrogenase from Streptococcus mutans: purification, characterization, and crossed antigenicity with ...
|
|
24437
|
Lactate dehydrogenase from Streptococcus mutans: purification, characterization, and crossed antigenicity with ...
|
|
24438
|
Lactate dehydrogenase from Streptococcus mutans: purification, characterization, and crossed antigenicity with ...
|
|
24439
|
Lactate dehydrogenase from Streptococcus mutans: purification, characterization, and crossed antigenicity with ...
|
|
24440
|
Lactate dehydrogenase from Streptococcus mutans: purification, characterization, and crossed antigenicity with ...
|
|
24441
|
Lactate dehydrogenase from Streptococcus mutans: purification, characterization, and crossed antigenicity with ...
|
|
24442
|
Lactate dehydrogenase from Streptococcus mutans: purification, characterization, and crossed antigenicity with ...
|
|
24443
|
Lactate dehydrogenase from Streptococcus mutans: purification, characterization, and crossed antigenicity with ...
|
|
24444
|
Lactate dehydrogenase from Streptococcus mutans: purification, characterization, and crossed antigenicity with ...
|
|
24445
|
Lactate dehydrogenase from Streptococcus mutans: purification, characterization, and crossed antigenicity with ...
|
|
24446
|
Lactate dehydrogenase from Streptococcus mutans: purification, characterization, and crossed antigenicity with ...
|
|
24447
|
Lactate dehydrogenase from Streptococcus mutans: purification, characterization, and crossed antigenicity with ...
|
|
24448
|
Lactate dehydrogenase from Streptococcus mutans: purification, characterization, and crossed antigenicity with ...
|
|
24449
|
Lactate dehydrogenase from Streptococcus mutans: purification, characterization, and crossed antigenicity with ...
|
|
24450
|
Purification and properties of Streptococcus mutans extracellular glucosyltransferase
|
|
24451
|
Purification and properties of Streptococcus mutans extracellular glucosyltransferase
|
|
24452
|
Purification and properties of Streptococcus mutans extracellular glucosyltransferase
|
|
24453
|
Structure-Activity Relationships and Thermal Stability of Human Glutathione Transferase P1-1 Governd by the ...
|
|
24454
|
Structure-Activity Relationships and Thermal Stability of Human Glutathione Transferase P1-1 Governd by the ...
|
|
24455
|
Structure-Activity Relationships and Thermal Stability of Human Glutathione Transferase P1-1 Governd by the ...
|
|
24456
|
Structure-Activity Relationships and Thermal Stability of Human Glutathione Transferase P1-1 Governd by the ...
|
|
24457
|
Structure-Activity Relationships and Thermal Stability of Human Glutathione Transferase P1-1 Governd by the ...
|
|
24458
|
Structure-Activity Relationships and Thermal Stability of Human Glutathione Transferase P1-1 Governd by the ...
|
|
24459
|
Structure-Activity Relationships and Thermal Stability of Human Glutathione Transferase P1-1 Governd by the ...
|
|
24460
|
Structure-Activity Relationships and Thermal Stability of Human Glutathione Transferase P1-1 Governd by the ...
|
|
24461
|
Structure-Activity Relationships and Thermal Stability of Human Glutathione Transferase P1-1 Governd by the ...
|
|
24462
|
Structure-Activity Relationships and Thermal Stability of Human Glutathione Transferase P1-1 Governd by the ...
|
|
24463
|
Structure-Activity Relationships and Thermal Stability of Human Glutathione Transferase P1-1 Governd by the ...
|
|
24464
|
Structure-Activity Relationships and Thermal Stability of Human Glutathione Transferase P1-1 Governd by the ...
|
|
24465
|
Structure-Activity Relationships and Thermal Stability of Human Glutathione Transferase P1-1 Governd by the ...
|
|
24466
|
Structure-Activity Relationships and Thermal Stability of Human Glutathione Transferase P1-1 Governd by the ...
|
|
24467
|
Structure-Activity Relationships and Thermal Stability of Human Glutathione Transferase P1-1 Governd by the ...
|
|
24468
|
Structure-Activity Relationships and Thermal Stability of Human Glutathione Transferase P1-1 Governd by the ...
|
|
24469
|
Structure-Activity Relationships and Thermal Stability of Human Glutathione Transferase P1-1 Governd by the ...
|
|
24470
|
Structure-Activity Relationships and Thermal Stability of Human Glutathione Transferase P1-1 Governd by the ...
|
|
24471
|
Structure-Activity Relationships and Thermal Stability of Human Glutathione Transferase P1-1 Governd by the ...
|
|
24472
|
Structure-Activity Relationships and Thermal Stability of Human Glutathione Transferase P1-1 Governd by the ...
|
|
24473
|
Structure-Activity Relationships and Thermal Stability of Human Glutathione Transferase P1-1 Governd by the ...
|
|
24474
|
Structure-Activity Relationships and Thermal Stability of Human Glutathione Transferase P1-1 Governd by the ...
|
|
24475
|
Structure-Activity Relationships and Thermal Stability of Human Glutathione Transferase P1-1 Governd by the ...
|
|
24476
|
Structure-Activity Relationships and Thermal Stability of Human Glutathione Transferase P1-1 Governd by the ...
|
|
24477
|
Isolation, characterization, and inhibition kinetics of enolase from Streptococcus rattus FA-1
|
|
24478
|
Isolation, characterization, and inhibition kinetics of enolase from Streptococcus rattus FA-1
|
|
24479
|
Isolation, characterization, and inhibition kinetics of enolase from Streptococcus rattus FA-1
|
|
24480
|
Isolation, characterization, and inhibition kinetics of enolase from Streptococcus rattus FA-1
|
|
24481
|
Purification and properties of pyruvate kinase from Streptococcus mutans
|
|
24482
|
Purification and properties of pyruvate kinase from Streptococcus mutans
|
|
24483
|
Purification and properties of pyruvate kinase from Streptococcus mutans
|
|
24484
|
Purification and properties of pyruvate kinase from Streptococcus mutans
|
|
24485
|
Purification and properties of pyruvate kinase from Streptococcus mutans
|
|
24486
|
Purification and properties of pyruvate kinase from Streptococcus mutans
|
|
24487
|
Purification and properties of pyruvate kinase from Streptococcus mutans
|
|
24488
|
Purification and properties of pyruvate kinase from Streptococcus mutans
|
|
24489
|
Purification and properties of pyruvate kinase from Streptococcus mutans
|
|
24490
|
Purification and properties of pyruvate kinase from Streptococcus mutans
|
|
24491
|
Purification and properties of pyruvate kinase from Streptococcus mutans
|
|
24492
|
Purification and properties of pyruvate kinase from Streptococcus mutans
|
|
24493
|
Purification and properties of pyruvate kinase from Streptococcus mutans
|
|
24494
|
Purification and properties of pyruvate kinase from Streptococcus mutans
|
|
24495
|
Purification and properties of pyruvate kinase from Streptococcus mutans
|
|
24496
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24497
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24498
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24499
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24500
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24501
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24502
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24503
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24504
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24505
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24506
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24507
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24508
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24509
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24510
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24511
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24512
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24513
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24514
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24515
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24516
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24517
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24518
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24519
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24520
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24521
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24522
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24523
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24524
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24525
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24526
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24527
|
Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium ...
|
|
24528
|
Effects of pH and energy supply on activity and amount of pyruvate formate-lyase in Streptococcus bovis
|
|
24529
|
Purification and properties of pyruvate kinase from Streptococcus sanguis and activator specificity of ...
|
|
24530
|
Purification and properties of pyruvate kinase from Streptococcus sanguis and activator specificity of ...
|
|
24531
|
Purification and properties of pyruvate kinase from Streptococcus sanguis and activator specificity of ...
|
|
24532
|
Purification and properties of pyruvate kinase from Streptococcus sanguis and activator specificity of ...
|
|
24533
|
Purification and properties of pyruvate kinase from Streptococcus sanguis and activator specificity of ...
|
|
24534
|
Purification and properties of pyruvate kinase from Streptococcus sanguis and activator specificity of ...
|
|
24535
|
Purification and properties of pyruvate kinase from Streptococcus sanguis and activator specificity of ...
|
|
24536
|
Purification and properties of pyruvate kinase from Streptococcus sanguis and activator specificity of ...
|
|
24537
|
Purification and properties of pyruvate kinase from Streptococcus sanguis and activator specificity of ...
|
|
24538
|
Purification and properties of pyruvate kinase from Streptococcus sanguis and activator specificity of ...
|
|
24539
|
Lactic acid translocation: terminal step in glycolysis by Streptococcus faecalis
|
|
24540
|
Lactic acid translocation: terminal step in glycolysis by Streptococcus faecalis
|
|
24541
|
Lactic acid translocation: terminal step in glycolysis by Streptococcus faecalis
|
|
24542
|
Purification and properties of dextransucrase from Streptococcus mutans
|
|
24543
|
Purification and properties of dextransucrase from Streptococcus mutans
|
|
24544
|
Exploring the Role of the Active Site Cysteine in Human Muscle Creatine Kinase
|
|
24545
|
Exploring the Role of the Active Site Cysteine in Human Muscle Creatine Kinase
|
|
24546
|
Exploring the Role of the Active Site Cysteine in Human Muscle Creatine Kinase
|
|
24547
|
Exploring the Role of the Active Site Cysteine in Human Muscle Creatine Kinase
|
|
24548
|
Exploring the Role of the Active Site Cysteine in Human Muscle Creatine Kinase
|
|
24549
|
Exploring the Role of the Active Site Cysteine in Human Muscle Creatine Kinase
|
|
24550
|
Exploring the Role of the Active Site Cysteine in Human Muscle Creatine Kinase
|
|
24551
|
Exploring the Role of the Active Site Cysteine in Human Muscle Creatine Kinase
|
|
24552
|
Exploring the Role of the Active Site Cysteine in Human Muscle Creatine Kinase
|
|
24553
|
Exploring the Role of the Active Site Cysteine in Human Muscle Creatine Kinase
|
|
24554
|
Identification of the putrescine biosynthetic genes in Pseudomonas aeruginosa and characterization of agmatine ...
|
|
24555
|
Identification of the putrescine biosynthetic genes in Pseudomonas aeruginosa and characterization of agmatine ...
|
|
24556
|
Purification and properties of rabbit heart muscle aldolase
|
|
24557
|
Mechanism-based inactivation of thioredoxin reductase from Plasmodium falciparum by Mannich bases. Implication ...
|
|
24558
|
Mechanism-based inactivation of thioredoxin reductase from Plasmodium falciparum by Mannich bases. Implication ...
|
|
24559
|
Mechanism-based inactivation of thioredoxin reductase from Plasmodium falciparum by Mannich bases. Implication ...
|
|
24560
|
Mechanism-based inactivation of thioredoxin reductase from Plasmodium falciparum by Mannich bases. Implication ...
|
|
24561
|
Mechanism-based inactivation of thioredoxin reductase from Plasmodium falciparum by Mannich bases. Implication ...
|
|
24562
|
Mechanism-based inactivation of thioredoxin reductase from Plasmodium falciparum by Mannich bases. Implication ...
|
|
24563
|
Mechanism-based inactivation of thioredoxin reductase from Plasmodium falciparum by Mannich bases. Implication ...
|
|
24564
|
Mechanism-based inactivation of thioredoxin reductase from Plasmodium falciparum by Mannich bases. Implication ...
|
|
24565
|
Mechanism-based inactivation of thioredoxin reductase from Plasmodium falciparum by Mannich bases. Implication ...
|
|
24566
|
Mechanism-based inactivation of thioredoxin reductase from Plasmodium falciparum by Mannich bases. Implication ...
|
|
24567
|
Mechanism-based inactivation of thioredoxin reductase from Plasmodium falciparum by Mannich bases. Implication ...
|
|
24568
|
Glyoxalase II from mouse liver
|
|
24569
|
Glyoxalase II from mouse liver
|
|
24570
|
Purification and characterization of NADH oxidase from Serpulina (Treponema) hyodysenteriae
|
|
24571
|
Molecular and biochemical characterization of 2-hydroxyisoflavanone dehydratase. Involvement of ...
|
|
24572
|
Molecular and biochemical characterization of 2-hydroxyisoflavanone dehydratase. Involvement of ...
|
|
24573
|
Molecular and biochemical characterization of 2-hydroxyisoflavanone dehydratase. Involvement of ...
|
|
24574
|
Molecular and biochemical characterization of 2-hydroxyisoflavanone dehydratase. Involvement of ...
|
|
24575
|
Molecular and biochemical characterization of 2-hydroxyisoflavanone dehydratase. Involvement of ...
|
|
24576
|
Molecular and biochemical characterization of 2-hydroxyisoflavanone dehydratase. Involvement of ...
|
|
24577
|
Glyoxysomal acetoacetyl-CoA thiolase and 3-oxoacyl-CoA thiolase from sunflower cotyledons
|
|
24578
|
Glyoxysomal acetoacetyl-CoA thiolase and 3-oxoacyl-CoA thiolase from sunflower cotyledons
|
|
24579
|
Glyoxysomal acetoacetyl-CoA thiolase and 3-oxoacyl-CoA thiolase from sunflower cotyledons
|
|
24580
|
Glyoxysomal acetoacetyl-CoA thiolase and 3-oxoacyl-CoA thiolase from sunflower cotyledons
|
|
24581
|
Glyoxysomal acetoacetyl-CoA thiolase and 3-oxoacyl-CoA thiolase from sunflower cotyledons
|
|
24582
|
Glyoxysomal acetoacetyl-CoA thiolase and 3-oxoacyl-CoA thiolase from sunflower cotyledons
|
|
24583
|
A Streptomyces collinus thiolase with novel acetyl-CoA:acyl carrier protein transacylase activity
|
|
24584
|
A Streptomyces collinus thiolase with novel acetyl-CoA:acyl carrier protein transacylase activity
|
|
24585
|
A Streptomyces collinus thiolase with novel acetyl-CoA:acyl carrier protein transacylase activity
|
|
24586
|
A Streptomyces collinus thiolase with novel acetyl-CoA:acyl carrier protein transacylase activity
|
|
24587
|
A Streptomyces collinus thiolase with novel acetyl-CoA:acyl carrier protein transacylase activity
|
|
24588
|
A Streptomyces collinus thiolase with novel acetyl-CoA:acyl carrier protein transacylase activity
|
|
24589
|
A Streptomyces collinus thiolase with novel acetyl-CoA:acyl carrier protein transacylase activity
|
|
24590
|
A Streptomyces collinus thiolase with novel acetyl-CoA:acyl carrier protein transacylase activity
|
|
24591
|
A Streptomyces collinus thiolase with novel acetyl-CoA:acyl carrier protein transacylase activity
|
|
24592
|
A Streptomyces collinus thiolase with novel acetyl-CoA:acyl carrier protein transacylase activity
|
|
24593
|
A Streptomyces collinus thiolase with novel acetyl-CoA:acyl carrier protein transacylase activity
|
|
24594
|
A Streptomyces collinus thiolase with novel acetyl-CoA:acyl carrier protein transacylase activity
|
|
24595
|
Mutational analysis of the active-site residues crucial for catalytic activity of adenosine kinase from ...
|
|
24596
|
Mutational analysis of the active-site residues crucial for catalytic activity of adenosine kinase from ...
|
|
24597
|
Mutational analysis of the active-site residues crucial for catalytic activity of adenosine kinase from ...
|
|
24598
|
Mutational analysis of the active-site residues crucial for catalytic activity of adenosine kinase from ...
|
|
24599
|
Mutational analysis of the active-site residues crucial for catalytic activity of adenosine kinase from ...
|
|
24600
|
Mutational analysis of the active-site residues crucial for catalytic activity of adenosine kinase from ...
|
|
24601
|
Mutational analysis of the active-site residues crucial for catalytic activity of adenosine kinase from ...
|
|
24602
|
Mutational analysis of the active-site residues crucial for catalytic activity of adenosine kinase from ...
|
|
24603
|
Mutational analysis of the active-site residues crucial for catalytic activity of adenosine kinase from ...
|
|
24604
|
Mutational analysis of the active-site residues crucial for catalytic activity of adenosine kinase from ...
|
|
24605
|
Mutational analysis of the active-site residues crucial for catalytic activity of adenosine kinase from ...
|
|
24606
|
Purification and properties of an alpha-methylacyl-CoA racemase from rat liver
|
|
24607
|
Purification and properties of an alpha-methylacyl-CoA racemase from rat liver
|
|
24608
|
Purification and properties of an alpha-methylacyl-CoA racemase from rat liver
|
|
24609
|
Purification and properties of an alpha-methylacyl-CoA racemase from rat liver
|
|
24610
|
Purification and properties of an alpha-methylacyl-CoA racemase from rat liver
|
|
24611
|
Purification and properties of an alpha-methylacyl-CoA racemase from rat liver
|
|
24612
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24613
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24614
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24615
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24616
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24617
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24618
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24619
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24620
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24621
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24622
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24623
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24624
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24625
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24626
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24627
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24628
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24629
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24630
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24631
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24632
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24633
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24634
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24635
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24636
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24637
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24638
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24639
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24640
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24641
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24642
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24643
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24644
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24645
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24646
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24647
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24648
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24649
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24650
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24651
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24652
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24653
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24654
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24655
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24656
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24657
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24658
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24659
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24660
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24661
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24662
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24663
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24664
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24665
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24666
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24667
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24668
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24669
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24670
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24671
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24672
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24673
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24674
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24675
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24676
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24677
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24678
|
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase
|
|
24679
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24680
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24681
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24682
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24683
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24684
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24685
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24686
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24687
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24688
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24689
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24690
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24691
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24692
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24693
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24694
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24695
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24696
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24697
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24698
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24699
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24700
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24701
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24702
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24703
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24704
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24705
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24706
|
Site-directed mutagenesis of beta-lactamase I. Single and double mutants of Glu-166 and Lys-73
|
|
24707
|
Isolation and analysis of mutated histidyl-tRNA synthetases from Escherichia coli
|
|
24708
|
Isolation and analysis of mutated histidyl-tRNA synthetases from Escherichia coli
|
|
24709
|
Isolation and analysis of mutated histidyl-tRNA synthetases from Escherichia coli
|
|
24710
|
Isolation and analysis of mutated histidyl-tRNA synthetases from Escherichia coli
|
|
24711
|
Isolation and analysis of mutated histidyl-tRNA synthetases from Escherichia coli
|
|
24712
|
Isolation and analysis of mutated histidyl-tRNA synthetases from Escherichia coli
|
|
24713
|
Isolation and analysis of mutated histidyl-tRNA synthetases from Escherichia coli
|
|
24714
|
Isolation and analysis of mutated histidyl-tRNA synthetases from Escherichia coli
|
|
24715
|
Isolation and analysis of mutated histidyl-tRNA synthetases from Escherichia coli
|
|
24716
|
Isolation and analysis of mutated histidyl-tRNA synthetases from Escherichia coli
|
|
24717
|
Isolation and analysis of mutated histidyl-tRNA synthetases from Escherichia coli
|
|
24718
|
Isolation and analysis of mutated histidyl-tRNA synthetases from Escherichia coli
|
|
24719
|
Isolation and analysis of mutated histidyl-tRNA synthetases from Escherichia coli
|
|
24720
|
Immunological and structural relatedness of isozymes and genetic variants of 3-phosphoglycerate kinase from ...
|
|
24721
|
Immunological and structural relatedness of isozymes and genetic variants of 3-phosphoglycerate kinase from ...
|
|
24722
|
Immunological and structural relatedness of isozymes and genetic variants of 3-phosphoglycerate kinase from ...
|
|
24723
|
Immunological and structural relatedness of isozymes and genetic variants of 3-phosphoglycerate kinase from ...
|
|
24724
|
Immunological and structural relatedness of isozymes and genetic variants of 3-phosphoglycerate kinase from ...
|
|
24725
|
Immunological and structural relatedness of isozymes and genetic variants of 3-phosphoglycerate kinase from ...
|
|
24726
|
Immunological and structural relatedness of isozymes and genetic variants of 3-phosphoglycerate kinase from ...
|
|
24727
|
Immunological and structural relatedness of isozymes and genetic variants of 3-phosphoglycerate kinase from ...
|
|
24728
|
Immunological and structural relatedness of isozymes and genetic variants of 3-phosphoglycerate kinase from ...
|
|
24729
|
Immunological and structural relatedness of isozymes and genetic variants of 3-phosphoglycerate kinase from ...
|
|
24730
|
Immunological and structural relatedness of isozymes and genetic variants of 3-phosphoglycerate kinase from ...
|
|
24731
|
Glucose-6-phosphate-dependent pyruvate kinase in Streptococcus mutans
|
|
24732
|
Glucose-6-phosphate-dependent pyruvate kinase in Streptococcus mutans
|
|
24733
|
Glucose-6-phosphate-dependent pyruvate kinase in Streptococcus mutans
|
|
24734
|
Glucose-6-phosphate-dependent pyruvate kinase in Streptococcus mutans
|
|
24741
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24742
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24743
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24744
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24745
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24746
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24747
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24748
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24749
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24750
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24751
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24752
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24753
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24754
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24755
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24756
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24757
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24758
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24759
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24760
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24761
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24762
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24763
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24764
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24765
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24766
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24767
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24768
|
Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and ...
|
|
24769
|
Effect of different NADH oxidase levels on glucose metabolism by Lactococcus lactis: kinetics of intracellular ...
|
|
24770
|
Glycolysis and the regulation of glucose transport in Lactococcus lactis spp. lactis in batch and fed-batch ...
|
|
24771
|
Glycolysis and the regulation of glucose transport in Lactococcus lactis spp. lactis in batch and fed-batch ...
|
|
24772
|
The Streptococcus faecalis oxidases for reduced diphosphopyridine nucleotide. III. Isolation and properties of ...
|
|
24773
|
The Streptococcus faecalis oxidases for reduced diphosphopyridine nucleotide. III. Isolation and properties of ...
|
|
24774
|
The Streptococcus faecalis oxidases for reduced diphosphopyridine nucleotide. III. Isolation and properties of ...
|
|
24775
|
The Streptococcus faecalis oxidases for reduced diphosphopyridine nucleotide. III. Isolation and properties of ...
|
|
24776
|
A human brain L-3-hydroxyacyl-coenzyme A dehydrogenase is identical to an amyloid beta-peptide-binding protein ...
|
|
24777
|
A human brain L-3-hydroxyacyl-coenzyme A dehydrogenase is identical to an amyloid beta-peptide-binding protein ...
|
|
24778
|
A human brain L-3-hydroxyacyl-coenzyme A dehydrogenase is identical to an amyloid beta-peptide-binding protein ...
|
|
24779
|
A human brain L-3-hydroxyacyl-coenzyme A dehydrogenase is identical to an amyloid beta-peptide-binding protein ...
|
|
24780
|
A human brain L-3-hydroxyacyl-coenzyme A dehydrogenase is identical to an amyloid beta-peptide-binding protein ...
|
|
24781
|
A human brain L-3-hydroxyacyl-coenzyme A dehydrogenase is identical to an amyloid beta-peptide-binding protein ...
|
|
24782
|
Acetyl-coenzyme A deacylase activity in liver is not an artifact. Subcellular distribution and substrate ...
|
|
24783
|
Acetyl-coenzyme A deacylase activity in liver is not an artifact. Subcellular distribution and substrate ...
|
|
24784
|
Acetyl-coenzyme A deacylase activity in liver is not an artifact. Subcellular distribution and substrate ...
|
|
24785
|
Acetyl-coenzyme A deacylase activity in liver is not an artifact. Subcellular distribution and substrate ...
|
|
24786
|
Acetyl-coenzyme A deacylase activity in liver is not an artifact. Subcellular distribution and substrate ...
|
|
24787
|
Acetyl-coenzyme A deacylase activity in liver is not an artifact. Subcellular distribution and substrate ...
|
|
24788
|
Acetyl-coenzyme A deacylase activity in liver is not an artifact. Subcellular distribution and substrate ...
|
|
24789
|
Acetyl-coenzyme A deacylase activity in liver is not an artifact. Subcellular distribution and substrate ...
|
|
24790
|
The human PICD gene encodes a cytoplasmic and peroxisomal NADP(+)-dependent isocitrate dehydrogenase
|
|
24791
|
Purification and properties of two succinate semialdehyde dehydrogenases from human brain
|
|
24792
|
Purification and properties of two succinate semialdehyde dehydrogenases from human brain
|
|
24793
|
Purification and properties of two succinate semialdehyde dehydrogenases from human brain
|
|
24794
|
Purification and properties of two succinate semialdehyde dehydrogenases from human brain
|
|
24795
|
Purification and properties of two succinate semialdehyde dehydrogenases from human brain
|
|
24796
|
Purification and properties of two succinate semialdehyde dehydrogenases from human brain
|
|
24797
|
Purification and properties of two succinate semialdehyde dehydrogenases from human brain
|
|
24798
|
Purification and properties of two succinate semialdehyde dehydrogenases from human brain
|
|
24799
|
Rabbit lung indolethylamine N-methyltransferase. cDNA and gene cloning and characterization
|
|
24800
|
Rabbit lung indolethylamine N-methyltransferase. cDNA and gene cloning and characterization
|
|
24801
|
Rabbit lung indolethylamine N-methyltransferase. cDNA and gene cloning and characterization
|
|
24802
|
Rabbit lung indolethylamine N-methyltransferase. cDNA and gene cloning and characterization
|
|
24803
|
Rabbit lung indolethylamine N-methyltransferase. cDNA and gene cloning and characterization
|
|
24804
|
Rabbit lung indolethylamine N-methyltransferase. cDNA and gene cloning and characterization
|
|
24805
|
Rabbit lung indolethylamine N-methyltransferase. cDNA and gene cloning and characterization
|
|
24806
|
Rabbit lung indolethylamine N-methyltransferase. cDNA and gene cloning and characterization
|
|
24807
|
Rabbit lung indolethylamine N-methyltransferase. cDNA and gene cloning and characterization
|
|
24808
|
Rabbit lung indolethylamine N-methyltransferase. cDNA and gene cloning and characterization
|
|
24809
|
Rabbit lung indolethylamine N-methyltransferase. cDNA and gene cloning and characterization
|
|
24810
|
Rabbit lung indolethylamine N-methyltransferase. cDNA and gene cloning and characterization
|
|
24811
|
Rabbit lung indolethylamine N-methyltransferase. cDNA and gene cloning and characterization
|
|
24812
|
Rabbit lung indolethylamine N-methyltransferase. cDNA and gene cloning and characterization
|
|
24813
|
Creatine kinase: the reactive cysteine is required for synergism but is nonessential for catalysis
|
|
24814
|
Creatine kinase: the reactive cysteine is required for synergism but is nonessential for catalysis
|
|
24815
|
Creatine kinase: the reactive cysteine is required for synergism but is nonessential for catalysis
|
|
24816
|
Creatine kinase: the reactive cysteine is required for synergism but is nonessential for catalysis
|
|
24817
|
Creatine kinase: the reactive cysteine is required for synergism but is nonessential for catalysis
|
|
24818
|
Creatine kinase: the reactive cysteine is required for synergism but is nonessential for catalysis
|
|
24819
|
Creatine kinase: the reactive cysteine is required for synergism but is nonessential for catalysis
|
|
24820
|
Creatine kinase: the reactive cysteine is required for synergism but is nonessential for catalysis
|
|
24821
|
Creatine kinase: the reactive cysteine is required for synergism but is nonessential for catalysis
|
|
24822
|
Creatine kinase: the reactive cysteine is required for synergism but is nonessential for catalysis
|
|
24823
|
Creatine kinase: the reactive cysteine is required for synergism but is nonessential for catalysis
|
|
24824
|
Creatine kinase: the reactive cysteine is required for synergism but is nonessential for catalysis
|
|
24825
|
Creatine kinase: the reactive cysteine is required for synergism but is nonessential for catalysis
|
|
24826
|
Creatine kinase: the reactive cysteine is required for synergism but is nonessential for catalysis
|
|
24827
|
Creatine kinase: the reactive cysteine is required for synergism but is nonessential for catalysis
|
|
24828
|
Creatine kinase: the reactive cysteine is required for synergism but is nonessential for catalysis
|
|
24829
|
Creatine kinase: the reactive cysteine is required for synergism but is nonessential for catalysis
|
|
24830
|
Creatine kinase: the reactive cysteine is required for synergism but is nonessential for catalysis
|
|
24831
|
Creatine kinase: the reactive cysteine is required for synergism but is nonessential for catalysis
|
|
24832
|
Creatine kinase: the reactive cysteine is required for synergism but is nonessential for catalysis
|
|
24833
|
Creatine kinase: the reactive cysteine is required for synergism but is nonessential for catalysis
|
|
24834
|
Creatine kinase: the reactive cysteine is required for synergism but is nonessential for catalysis
|
|
24835
|
Kinetic characterization of active site mutants Ser402Ala and Phe397His of vanadium chloroperoxidase from the ...
|
|
24836
|
Kinetic characterization of active site mutants Ser402Ala and Phe397His of vanadium chloroperoxidase from the ...
|
|
24837
|
Kinetic characterization of active site mutants Ser402Ala and Phe397His of vanadium chloroperoxidase from the ...
|
|
24838
|
Kinetic characterization of active site mutants Ser402Ala and Phe397His of vanadium chloroperoxidase from the ...
|
|
24839
|
Kinetic characterization of active site mutants Ser402Ala and Phe397His of vanadium chloroperoxidase from the ...
|
|
24840
|
Kinetic characterization of active site mutants Ser402Ala and Phe397His of vanadium chloroperoxidase from the ...
|
|
24841
|
Kinetic characterization of active site mutants Ser402Ala and Phe397His of vanadium chloroperoxidase from the ...
|
|
24842
|
Kinetic characterization of active site mutants Ser402Ala and Phe397His of vanadium chloroperoxidase from the ...
|
|
24843
|
Kinetic characterization of active site mutants Ser402Ala and Phe397His of vanadium chloroperoxidase from the ...
|
|
24844
|
Kinetic characterization of active site mutants Ser402Ala and Phe397His of vanadium chloroperoxidase from the ...
|
|
24845
|
Kinetic characterization of active site mutants Ser402Ala and Phe397His of vanadium chloroperoxidase from the ...
|
|
24846
|
Kinetic characterization of active site mutants Ser402Ala and Phe397His of vanadium chloroperoxidase from the ...
|
|
24847
|
Kinetic characterization of active site mutants Ser402Ala and Phe397His of vanadium chloroperoxidase from the ...
|
|
24848
|
Kinetic characterization of active site mutants Ser402Ala and Phe397His of vanadium chloroperoxidase from the ...
|
|
24849
|
Kinetic characterization of active site mutants Ser402Ala and Phe397His of vanadium chloroperoxidase from the ...
|
|
24850
|
Kinetic characterization of active site mutants Ser402Ala and Phe397His of vanadium chloroperoxidase from the ...
|
|
24851
|
Differential contributions of NADPH-cytochrome P450 oxidoreductase FAD binding site residues to flavin binding ...
|
|
24852
|
Differential contributions of NADPH-cytochrome P450 oxidoreductase FAD binding site residues to flavin binding ...
|
|
24853
|
Differential contributions of NADPH-cytochrome P450 oxidoreductase FAD binding site residues to flavin binding ...
|
|
24854
|
Differential contributions of NADPH-cytochrome P450 oxidoreductase FAD binding site residues to flavin binding ...
|
|
24855
|
Differential contributions of NADPH-cytochrome P450 oxidoreductase FAD binding site residues to flavin binding ...
|
|
24856
|
Differential contributions of NADPH-cytochrome P450 oxidoreductase FAD binding site residues to flavin binding ...
|
|
24857
|
Differential contributions of NADPH-cytochrome P450 oxidoreductase FAD binding site residues to flavin binding ...
|
|
24858
|
Differential contributions of NADPH-cytochrome P450 oxidoreductase FAD binding site residues to flavin binding ...
|
|
24859
|
Differential contributions of NADPH-cytochrome P450 oxidoreductase FAD binding site residues to flavin binding ...
|
|
24860
|
Differential contributions of NADPH-cytochrome P450 oxidoreductase FAD binding site residues to flavin binding ...
|
|
24861
|
Differential contributions of NADPH-cytochrome P450 oxidoreductase FAD binding site residues to flavin binding ...
|
|
24862
|
Differential contributions of NADPH-cytochrome P450 oxidoreductase FAD binding site residues to flavin binding ...
|
|
24863
|
Differential contributions of NADPH-cytochrome P450 oxidoreductase FAD binding site residues to flavin binding ...
|
|
24864
|
Pig muscle aldehyde reductase. Identity of pig muscle aldehyde reductase with pig lens aldose reductase and ...
|
|
24865
|
Pig muscle aldehyde reductase. Identity of pig muscle aldehyde reductase with pig lens aldose reductase and ...
|
|
24866
|
Pig muscle aldehyde reductase. Identity of pig muscle aldehyde reductase with pig lens aldose reductase and ...
|
|
24867
|
Pig muscle aldehyde reductase. Identity of pig muscle aldehyde reductase with pig lens aldose reductase and ...
|
|
24868
|
Pig muscle aldehyde reductase. Identity of pig muscle aldehyde reductase with pig lens aldose reductase and ...
|
|
24869
|
Pig muscle aldehyde reductase. Identity of pig muscle aldehyde reductase with pig lens aldose reductase and ...
|
|
24870
|
Pig muscle aldehyde reductase. Identity of pig muscle aldehyde reductase with pig lens aldose reductase and ...
|
|
24871
|
Pig muscle aldehyde reductase. Identity of pig muscle aldehyde reductase with pig lens aldose reductase and ...
|
|
24872
|
Pig muscle aldehyde reductase. Identity of pig muscle aldehyde reductase with pig lens aldose reductase and ...
|
|
24873
|
Pig muscle aldehyde reductase. Identity of pig muscle aldehyde reductase with pig lens aldose reductase and ...
|
|
24874
|
Pig muscle aldehyde reductase. Identity of pig muscle aldehyde reductase with pig lens aldose reductase and ...
|
|
24875
|
Pig muscle aldehyde reductase. Identity of pig muscle aldehyde reductase with pig lens aldose reductase and ...
|
|
24876
|
Pig muscle aldehyde reductase. Identity of pig muscle aldehyde reductase with pig lens aldose reductase and ...
|
|
24877
|
Identification and characterization of the first class of potent bacterial acetyl-CoA carboxylase inhibitors ...
|
|
24878
|
Identification and characterization of the first class of potent bacterial acetyl-CoA carboxylase inhibitors ...
|
|
24879
|
Identification and characterization of the first class of potent bacterial acetyl-CoA carboxylase inhibitors ...
|
|
24880
|
Identification and characterization of the first class of potent bacterial acetyl-CoA carboxylase inhibitors ...
|
|
24881
|
Identification and characterization of the first class of potent bacterial acetyl-CoA carboxylase inhibitors ...
|
|
24882
|
Identification and characterization of the first class of potent bacterial acetyl-CoA carboxylase inhibitors ...
|
|
24883
|
Identification and characterization of the first class of potent bacterial acetyl-CoA carboxylase inhibitors ...
|
|
24884
|
Identification and characterization of the first class of potent bacterial acetyl-CoA carboxylase inhibitors ...
|
|
24885
|
Identification and characterization of the first class of potent bacterial acetyl-CoA carboxylase inhibitors ...
|
|
24886
|
Identification and characterization of the first class of potent bacterial acetyl-CoA carboxylase inhibitors ...
|
|
24887
|
Graphical analyses on binding of ligands to a two-sited system. Theoretical treatments exemplified on yeast ...
|
|
24888
|
Graphical analyses on binding of ligands to a two-sited system. Theoretical treatments exemplified on yeast ...
|
|
24889
|
Variation in the divalent cation requirements of influenza A virus N1 neuraminidases
|
|
24890
|
Variation in the divalent cation requirements of influenza A virus N1 neuraminidases
|
|
24891
|
Variation in the divalent cation requirements of influenza A virus N1 neuraminidases
|
|
24892
|
Variation in the divalent cation requirements of influenza A virus N1 neuraminidases
|
|
24893
|
Variation in the divalent cation requirements of influenza A virus N1 neuraminidases
|
|
24894
|
Variation in the divalent cation requirements of influenza A virus N1 neuraminidases
|
|
24895
|
Variation in the divalent cation requirements of influenza A virus N1 neuraminidases
|
|
24896
|
Variation in the divalent cation requirements of influenza A virus N1 neuraminidases
|
|
24897
|
Variation in the divalent cation requirements of influenza A virus N1 neuraminidases
|
|
24898
|
Variation in the divalent cation requirements of influenza A virus N1 neuraminidases
|
|
24899
|
Variation in the divalent cation requirements of influenza A virus N1 neuraminidases
|
|
24900
|
Variation in the divalent cation requirements of influenza A virus N1 neuraminidases
|
|
24901
|
Groups on the side chain of T252 in Escherichia coli leucyl-tRNA synthetase are important for discrimination ...
|
|
24902
|
Groups on the side chain of T252 in Escherichia coli leucyl-tRNA synthetase are important for discrimination ...
|
|
24903
|
Groups on the side chain of T252 in Escherichia coli leucyl-tRNA synthetase are important for discrimination ...
|
|
24904
|
Groups on the side chain of T252 in Escherichia coli leucyl-tRNA synthetase are important for discrimination ...
|
|
24905
|
Groups on the side chain of T252 in Escherichia coli leucyl-tRNA synthetase are important for discrimination ...
|
|
24906
|
Groups on the side chain of T252 in Escherichia coli leucyl-tRNA synthetase are important for discrimination ...
|
|
24907
|
Groups on the side chain of T252 in Escherichia coli leucyl-tRNA synthetase are important for discrimination ...
|
|
24908
|
Groups on the side chain of T252 in Escherichia coli leucyl-tRNA synthetase are important for discrimination ...
|
|
24909
|
Groups on the side chain of T252 in Escherichia coli leucyl-tRNA synthetase are important for discrimination ...
|
|
24910
|
The 2.2 A resolution structure of RpiB/AlsB from Escherichia coli illustrates a new approach to the ...
|
|
24911
|
The 2.2 A resolution structure of RpiB/AlsB from Escherichia coli illustrates a new approach to the ...
|
|
24912
|
The 2.2 A resolution structure of RpiB/AlsB from Escherichia coli illustrates a new approach to the ...
|
|
24913
|
The 2.2 A resolution structure of RpiB/AlsB from Escherichia coli illustrates a new approach to the ...
|
|
24914
|
Chicken ornithine transcarbamylase: purification and some properties
|
|
24915
|
Chicken ornithine transcarbamylase: purification and some properties
|
|
24916
|
Chicken ornithine transcarbamylase: purification and some properties
|
|
24917
|
Chicken ornithine transcarbamylase: purification and some properties
|
|
24918
|
Chicken ornithine transcarbamylase: purification and some properties
|
|
24919
|
Chicken ornithine transcarbamylase: purification and some properties
|
|
24920
|
Molecular abnormality of phosphoglycerate kinase-Uppsala associated with chronic nonspherocytic hemolytic ...
|
|
24921
|
Molecular abnormality of phosphoglycerate kinase-Uppsala associated with chronic nonspherocytic hemolytic ...
|
|
24922
|
Molecular abnormality of phosphoglycerate kinase-Uppsala associated with chronic nonspherocytic hemolytic ...
|
|
24923
|
Molecular abnormality of phosphoglycerate kinase-Uppsala associated with chronic nonspherocytic hemolytic ...
|
|
24924
|
Molecular abnormality of phosphoglycerate kinase-Uppsala associated with chronic nonspherocytic hemolytic ...
|
|
24925
|
Molecular abnormality of phosphoglycerate kinase-Uppsala associated with chronic nonspherocytic hemolytic ...
|
|
24926
|
Molecular abnormality of phosphoglycerate kinase-Uppsala associated with chronic nonspherocytic hemolytic ...
|
|
24927
|
Molecular abnormality of phosphoglycerate kinase-Uppsala associated with chronic nonspherocytic hemolytic ...
|
|
24928
|
A Thermoanaerobacter ethanolicus secondary alcohol dehydrogenase mutant derivative highly active and ...
|
|
24929
|
A Thermoanaerobacter ethanolicus secondary alcohol dehydrogenase mutant derivative highly active and ...
|
|
24930
|
A Thermoanaerobacter ethanolicus secondary alcohol dehydrogenase mutant derivative highly active and ...
|
|
24931
|
A Thermoanaerobacter ethanolicus secondary alcohol dehydrogenase mutant derivative highly active and ...
|
|
24932
|
A Thermoanaerobacter ethanolicus secondary alcohol dehydrogenase mutant derivative highly active and ...
|
|
24933
|
A Thermoanaerobacter ethanolicus secondary alcohol dehydrogenase mutant derivative highly active and ...
|
|
24934
|
Characterization of two ornithine carbamoyltransferases from Pseudomonas syringae pv. phaseolicola, the ...
|
|
24935
|
Characterization of two ornithine carbamoyltransferases from Pseudomonas syringae pv. phaseolicola, the ...
|
|
24936
|
Amphioxus alcohol dehydrogenase is a class 3 form of single type and of structural conservation but with ...
|
|
24937
|
Amphioxus alcohol dehydrogenase is a class 3 form of single type and of structural conservation but with ...
|
|
24938
|
Amphioxus alcohol dehydrogenase is a class 3 form of single type and of structural conservation but with ...
|
|
24939
|
Acetyl Coenzyme A Acetyltransferase of Rhizobium sp. (Cicer) Strain CC 1192
|
|
24940
|
Acetyl Coenzyme A Acetyltransferase of Rhizobium sp. (Cicer) Strain CC 1192
|
|
24941
|
Acetyl Coenzyme A Acetyltransferase of Rhizobium sp. (Cicer) Strain CC 1192
|
|
24942
|
Acetyl Coenzyme A Acetyltransferase of Rhizobium sp. (Cicer) Strain CC 1192
|
|
24943
|
Acetyl Coenzyme A Acetyltransferase of Rhizobium sp. (Cicer) Strain CC 1192
|
|
24944
|
Acetyl Coenzyme A Acetyltransferase of Rhizobium sp. (Cicer) Strain CC 1192
|
|
24945
|
Acetyl Coenzyme A Acetyltransferase of Rhizobium sp. (Cicer) Strain CC 1192
|
|
24946
|
Mutagenesis of histidinol dehydrogenase reveals roles for conserved histidine residues
|
|
24947
|
Mutagenesis of histidinol dehydrogenase reveals roles for conserved histidine residues
|
|
24948
|
Mutagenesis of histidinol dehydrogenase reveals roles for conserved histidine residues
|
|
24949
|
Mutagenesis of histidinol dehydrogenase reveals roles for conserved histidine residues
|
|
24950
|
Mutagenesis of histidinol dehydrogenase reveals roles for conserved histidine residues
|
|
24951
|
Mutagenesis of histidinol dehydrogenase reveals roles for conserved histidine residues
|
|
24952
|
Mutagenesis of histidinol dehydrogenase reveals roles for conserved histidine residues
|
|
24953
|
Mutagenesis of histidinol dehydrogenase reveals roles for conserved histidine residues
|
|
24954
|
Mutagenesis of histidinol dehydrogenase reveals roles for conserved histidine residues
|
|
24955
|
Mutagenesis of histidinol dehydrogenase reveals roles for conserved histidine residues
|
|
24956
|
Mutagenesis of histidinol dehydrogenase reveals roles for conserved histidine residues
|
|
24957
|
Mutagenesis of histidinol dehydrogenase reveals roles for conserved histidine residues
|
|
24958
|
Mutagenesis of histidinol dehydrogenase reveals roles for conserved histidine residues
|
|
24959
|
Mutagenesis of histidinol dehydrogenase reveals roles for conserved histidine residues
|
|
24960
|
Mutagenesis of histidinol dehydrogenase reveals roles for conserved histidine residues
|
|
24961
|
Mutagenesis of histidinol dehydrogenase reveals roles for conserved histidine residues
|
|
24962
|
Mutagenesis of histidinol dehydrogenase reveals roles for conserved histidine residues
|
|
24963
|
Fumarate reductase activity of Streptococcus faecalis
|
|
24964
|
Dihydrofolic reductase from Streptococcus faecalis R
|
|
24965
|
Dihydrofolic reductase from Streptococcus faecalis R
|
|
24966
|
Dihydrofolic reductase from Streptococcus faecalis R
|
|
24967
|
Dihydrofolic reductase from Streptococcus faecalis R
|
|
24968
|
Plant P5C reductase as a new target for aminomethylenebisphosphonates
|
|
24969
|
Plant P5C reductase as a new target for aminomethylenebisphosphonates
|
|
24970
|
Plant P5C reductase as a new target for aminomethylenebisphosphonates
|
|
24971
|
Plant P5C reductase as a new target for aminomethylenebisphosphonates
|
|
24972
|
Plant P5C reductase as a new target for aminomethylenebisphosphonates
|
|
24973
|
Plant P5C reductase as a new target for aminomethylenebisphosphonates
|
|
24974
|
Identification of two arginase isoenzyme activities along the nephron of Meriones shawi
|
|
24975
|
Identification of two arginase isoenzyme activities along the nephron of Meriones shawi
|
|
24976
|
Identification of two arginase isoenzyme activities along the nephron of Meriones shawi
|
|
24977
|
Identification of two arginase isoenzyme activities along the nephron of Meriones shawi
|
|
24978
|
Identification of two arginase isoenzyme activities along the nephron of Meriones shawi
|
|
24979
|
Combinatorial engineering to enhance amylosucrase performance: construction, selection, and screening of ...
|
|
24980
|
Combinatorial engineering to enhance amylosucrase performance: construction, selection, and screening of ...
|
|
24981
|
Combinatorial engineering to enhance amylosucrase performance: construction, selection, and screening of ...
|
|
24982
|
Combinatorial engineering to enhance amylosucrase performance: construction, selection, and screening of ...
|
|
24983
|
Combinatorial engineering to enhance amylosucrase performance: construction, selection, and screening of ...
|
|
24984
|
Combinatorial engineering to enhance amylosucrase performance: construction, selection, and screening of ...
|
|
24985
|
Combinatorial engineering to enhance amylosucrase performance: construction, selection, and screening of ...
|
|
24986
|
Combinatorial engineering to enhance amylosucrase performance: construction, selection, and screening of ...
|
|
24987
|
Combinatorial engineering to enhance amylosucrase performance: construction, selection, and screening of ...
|
|
24988
|
Combinatorial engineering to enhance amylosucrase performance: construction, selection, and screening of ...
|
|
24989
|
Combinatorial engineering to enhance amylosucrase performance: construction, selection, and screening of ...
|
|
24990
|
Combinatorial engineering to enhance amylosucrase performance: construction, selection, and screening of ...
|
|
24991
|
Studies on human beta-D-N-acetylhexosaminidases. II. Kinetic and structural properties
|
|
24992
|
Studies on human beta-D-N-acetylhexosaminidases. II. Kinetic and structural properties
|
|
24993
|
Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from ...
|
|
24994
|
Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from ...
|
|
24995
|
Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from ...
|
|
24996
|
Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from ...
|
|
24997
|
Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from ...
|
|
24998
|
Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from ...
|
|
24999
|
Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from ...
|
|
25000
|
Mutational analysis of the triclosan-binding region of enoyl-ACP (acyl-carrier protein) reductase from ...
|





 If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.
If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.