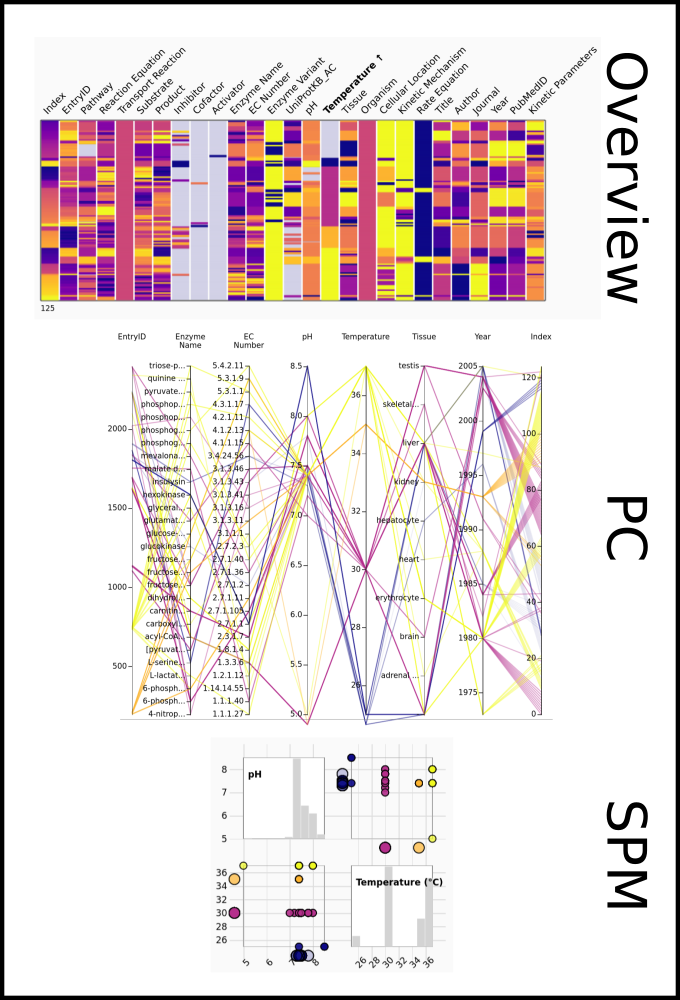
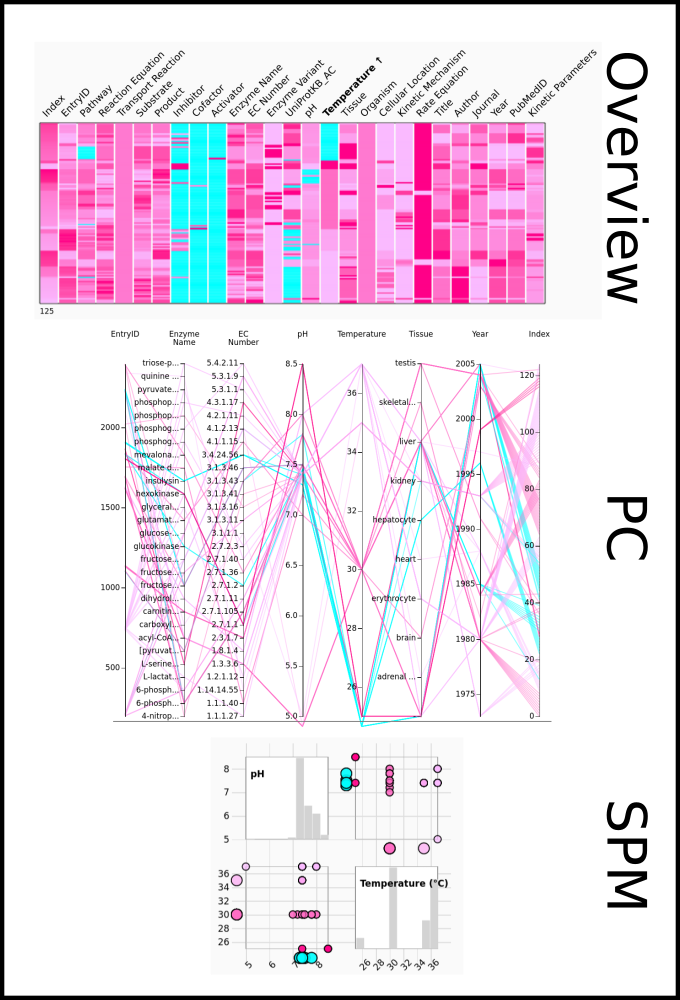
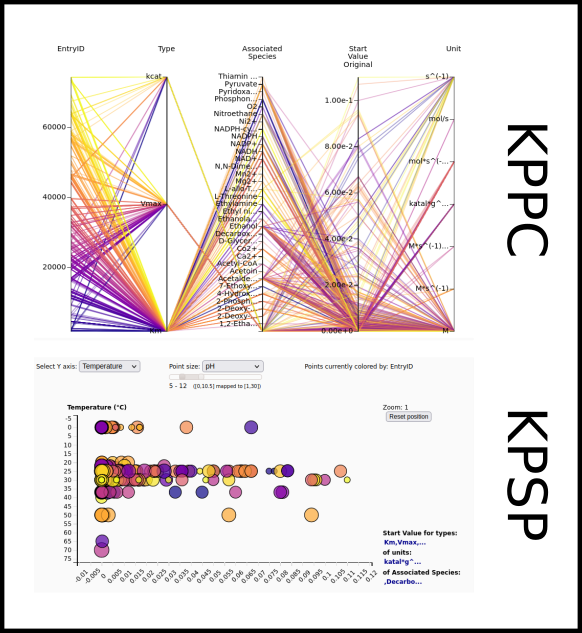
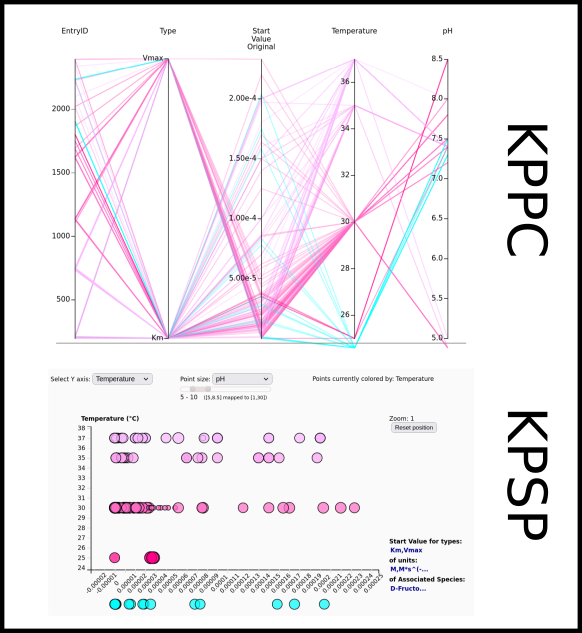
To improve SABIO-RK data interoperability, semantic markup was added to web pages as described and defined by
This structured information makes it easier to discover, collate and analyse our data.
|
14001
|
Steady-state kinetic analysis of the quinoprotein methylamine dehydrogenase from Paracoccus denitrificans
|
|
14002
|
Steady-state kinetic analysis of the quinoprotein methylamine dehydrogenase from Paracoccus denitrificans
|
|
14003
|
Steady-state kinetic analysis of the quinoprotein methylamine dehydrogenase from Paracoccus denitrificans
|
|
14004
|
Steady-state kinetic analysis of the quinoprotein methylamine dehydrogenase from Paracoccus denitrificans
|
|
14005
|
Steady-state kinetic analysis of the quinoprotein methylamine dehydrogenase from Paracoccus denitrificans
|
|
14006
|
Steady-state kinetic analysis of the quinoprotein methylamine dehydrogenase from Paracoccus denitrificans
|
|
14007
|
Steady-state kinetic analysis of the quinoprotein methylamine dehydrogenase from Paracoccus denitrificans
|
|
14008
|
Steady-state kinetic analysis of the quinoprotein methylamine dehydrogenase from Paracoccus denitrificans
|
|
14009
|
Steady-state kinetic analysis of the quinoprotein methylamine dehydrogenase from Paracoccus denitrificans
|
|
14010
|
Steady-state kinetic analysis of the quinoprotein methylamine dehydrogenase from Paracoccus denitrificans
|
|
14011
|
Steady-state kinetic analysis of the quinoprotein methylamine dehydrogenase from Paracoccus denitrificans
|
|
14012
|
Steady-state kinetic analysis of the quinoprotein methylamine dehydrogenase from Paracoccus denitrificans
|
|
14013
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14014
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14015
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14016
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14017
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14018
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14019
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14020
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14021
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14022
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14023
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14024
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14025
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14026
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14027
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14028
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14029
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14030
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14031
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14032
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14033
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14034
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14035
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14036
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14037
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14038
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14039
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14040
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14041
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14042
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14043
|
Overall Kinetic Mechanism of Saccharopine Dehydrogenase from Saccharomyces cerevisiae
|
|
14070
|
Metal ion specificity at the catalytic site of yeast enolase
|
|
14071
|
Metal ion specificity at the catalytic site of yeast enolase
|
|
14072
|
Metal ion specificity at the catalytic site of yeast enolase
|
|
14073
|
Metal ion specificity at the catalytic site of yeast enolase
|
|
14074
|
Metal ion specificity at the catalytic site of yeast enolase
|
|
14075
|
Metal ion specificity at the catalytic site of yeast enolase
|
|
14076
|
Metal ion specificity at the catalytic site of yeast enolase
|
|
14077
|
Metal ion specificity at the catalytic site of yeast enolase
|
|
14078
|
Metal ion specificity at the catalytic site of yeast enolase
|
|
14079
|
Metal ion specificity at the catalytic site of yeast enolase
|
|
14080
|
Metal ion specificity at the catalytic site of yeast enolase
|
|
14081
|
Metal ion specificity at the catalytic site of yeast enolase
|
|
14082
|
Metal ion specificity at the catalytic site of yeast enolase
|
|
14083
|
Metal ion specificity at the catalytic site of yeast enolase
|
|
14084
|
Metal ion specificity at the catalytic site of yeast enolase
|
|
14085
|
Metal ion specificity at the catalytic site of yeast enolase
|
|
14086
|
Metal ion specificity at the catalytic site of yeast enolase
|
|
14087
|
Methylthioadenosine phosphorylase from the archaeon Pyrococcus furiosus. Mechanism of the reaction and ...
|
|
14088
|
Methylthioadenosine phosphorylase from the archaeon Pyrococcus furiosus. Mechanism of the reaction and ...
|
|
14089
|
Methylthioadenosine phosphorylase from the archaeon Pyrococcus furiosus. Mechanism of the reaction and ...
|
|
14090
|
Methylthioadenosine phosphorylase from the archaeon Pyrococcus furiosus. Mechanism of the reaction and ...
|
|
14091
|
Methylthioadenosine phosphorylase from the archaeon Pyrococcus furiosus. Mechanism of the reaction and ...
|
|
14092
|
Methylthioadenosine phosphorylase from the archaeon Pyrococcus furiosus. Mechanism of the reaction and ...
|
|
14093
|
Methylthioadenosine phosphorylase from the archaeon Pyrococcus furiosus. Mechanism of the reaction and ...
|
|
14094
|
Methylthioadenosine phosphorylase from the archaeon Pyrococcus furiosus. Mechanism of the reaction and ...
|
|
14095
|
Methylthioadenosine phosphorylase from the archaeon Pyrococcus furiosus. Mechanism of the reaction and ...
|
|
14096
|
Pathway for the synthesis of mannosylglycerate in the hyperthermophilic archaeon Pyrococcus horikoshii. ...
|
|
14097
|
Pathway for the synthesis of mannosylglycerate in the hyperthermophilic archaeon Pyrococcus horikoshii. ...
|
|
14098
|
Pathway for the synthesis of mannosylglycerate in the hyperthermophilic archaeon Pyrococcus horikoshii. ...
|
|
14099
|
Pathway for the synthesis of mannosylglycerate in the hyperthermophilic archaeon Pyrococcus horikoshii. ...
|
|
14100
|
Pathway for the synthesis of mannosylglycerate in the hyperthermophilic archaeon Pyrococcus horikoshii. ...
|
|
14101
|
Pathway for the synthesis of mannosylglycerate in the hyperthermophilic archaeon Pyrococcus horikoshii. ...
|
|
14102
|
Pathway for the synthesis of mannosylglycerate in the hyperthermophilic archaeon Pyrococcus horikoshii. ...
|
|
14103
|
Pathway for the synthesis of mannosylglycerate in the hyperthermophilic archaeon Pyrococcus horikoshii. ...
|
|
14104
|
Pathway for the synthesis of mannosylglycerate in the hyperthermophilic archaeon Pyrococcus horikoshii. ...
|
|
14105
|
Pathway for the synthesis of mannosylglycerate in the hyperthermophilic archaeon Pyrococcus horikoshii. ...
|
|
14106
|
Pathway for the synthesis of mannosylglycerate in the hyperthermophilic archaeon Pyrococcus horikoshii. ...
|
|
14107
|
Pathway for the synthesis of mannosylglycerate in the hyperthermophilic archaeon Pyrococcus horikoshii. ...
|
|
14108
|
Characterization of human UDP-glucose dehydrogenase. CYS-276 is required for the second of two successive ...
|
|
14109
|
Characterization of human UDP-glucose dehydrogenase. CYS-276 is required for the second of two successive ...
|
|
14110
|
Characterization of human UDP-glucose dehydrogenase. CYS-276 is required for the second of two successive ...
|
|
14111
|
Characterization of human UDP-glucose dehydrogenase. CYS-276 is required for the second of two successive ...
|
|
14112
|
Characterization of human UDP-glucose dehydrogenase. CYS-276 is required for the second of two successive ...
|
|
14113
|
Site-directed mutagenesis of a hexapeptide segment involved in substrate recognition of phenylalanine ...
|
|
14114
|
Site-directed mutagenesis of a hexapeptide segment involved in substrate recognition of phenylalanine ...
|
|
14115
|
Site-directed mutagenesis of a hexapeptide segment involved in substrate recognition of phenylalanine ...
|
|
14116
|
Site-directed mutagenesis of a hexapeptide segment involved in substrate recognition of phenylalanine ...
|
|
14117
|
Site-directed mutagenesis of a hexapeptide segment involved in substrate recognition of phenylalanine ...
|
|
14118
|
Site-directed mutagenesis of a hexapeptide segment involved in substrate recognition of phenylalanine ...
|
|
14119
|
Site-directed mutagenesis of a hexapeptide segment involved in substrate recognition of phenylalanine ...
|
|
14120
|
Site-directed mutagenesis of a hexapeptide segment involved in substrate recognition of phenylalanine ...
|
|
14121
|
Site-directed mutagenesis of a hexapeptide segment involved in substrate recognition of phenylalanine ...
|
|
14122
|
Site-directed mutagenesis of a hexapeptide segment involved in substrate recognition of phenylalanine ...
|
|
14123
|
Site-directed mutagenesis of a hexapeptide segment involved in substrate recognition of phenylalanine ...
|
|
14124
|
Site-directed mutagenesis of a hexapeptide segment involved in substrate recognition of phenylalanine ...
|
|
14125
|
Site-directed mutagenesis of a hexapeptide segment involved in substrate recognition of phenylalanine ...
|
|
14126
|
Site-directed mutagenesis of a hexapeptide segment involved in substrate recognition of phenylalanine ...
|
|
14127
|
Site-directed mutagenesis of a hexapeptide segment involved in substrate recognition of phenylalanine ...
|
|
14128
|
Site-directed mutagenesis of a hexapeptide segment involved in substrate recognition of phenylalanine ...
|
|
14129
|
Site-directed mutagenesis of a hexapeptide segment involved in substrate recognition of phenylalanine ...
|
|
14130
|
Site-directed mutagenesis of a hexapeptide segment involved in substrate recognition of phenylalanine ...
|
|
14131
|
Site-directed mutagenesis of a hexapeptide segment involved in substrate recognition of phenylalanine ...
|
|
14132
|
Site-directed mutagenesis of a hexapeptide segment involved in substrate recognition of phenylalanine ...
|
|
14133
|
Site-directed mutagenesis of a hexapeptide segment involved in substrate recognition of phenylalanine ...
|
|
14134
|
Site-directed mutagenesis of a hexapeptide segment involved in substrate recognition of phenylalanine ...
|
|
14135
|
Site-directed mutagenesis of a hexapeptide segment involved in substrate recognition of phenylalanine ...
|
|
14136
|
Site-directed mutagenesis of a hexapeptide segment involved in substrate recognition of phenylalanine ...
|
|
14137
|
Site-directed mutagenesis of a hexapeptide segment involved in substrate recognition of phenylalanine ...
|
|
14138
|
Site-directed mutagenesis of a hexapeptide segment involved in substrate recognition of phenylalanine ...
|
|
14139
|
Insulin does not change the intracellular distribution of hexokinase in rat heart
|
|
14140
|
Insulin does not change the intracellular distribution of hexokinase in rat heart
|
|
14141
|
Insulin does not change the intracellular distribution of hexokinase in rat heart
|
|
14142
|
Insulin does not change the intracellular distribution of hexokinase in rat heart
|
|
14143
|
Insulin does not change the intracellular distribution of hexokinase in rat heart
|
|
14144
|
Insulin does not change the intracellular distribution of hexokinase in rat heart
|
|
14145
|
Insulin does not change the intracellular distribution of hexokinase in rat heart
|
|
14146
|
Insulin does not change the intracellular distribution of hexokinase in rat heart
|
|
14147
|
Insulin does not change the intracellular distribution of hexokinase in rat heart
|
|
14148
|
Insulin does not change the intracellular distribution of hexokinase in rat heart
|
|
14149
|
Insulin does not change the intracellular distribution of hexokinase in rat heart
|
|
14150
|
Insulin does not change the intracellular distribution of hexokinase in rat heart
|
|
14151
|
Insulin does not change the intracellular distribution of hexokinase in rat heart
|
|
14152
|
Insulin does not change the intracellular distribution of hexokinase in rat heart
|
|
14153
|
Insulin does not change the intracellular distribution of hexokinase in rat heart
|
|
14154
|
Insulin does not change the intracellular distribution of hexokinase in rat heart
|
|
14155
|
Insulin does not change the intracellular distribution of hexokinase in rat heart
|
|
14156
|
Insulin does not change the intracellular distribution of hexokinase in rat heart
|
|
14157
|
Insulin does not change the intracellular distribution of hexokinase in rat heart
|
|
14158
|
Insulin does not change the intracellular distribution of hexokinase in rat heart
|
|
14159
|
The fast-reacting sulfhydryl group of rat muscle phosphoglycerate kinase is necessary for activity and ...
|
|
14160
|
The fast-reacting sulfhydryl group of rat muscle phosphoglycerate kinase is necessary for activity and ...
|
|
14161
|
The fast-reacting sulfhydryl group of rat muscle phosphoglycerate kinase is necessary for activity and ...
|
|
14162
|
The fast-reacting sulfhydryl group of rat muscle phosphoglycerate kinase is necessary for activity and ...
|
|
14163
|
The fast-reacting sulfhydryl group of rat muscle phosphoglycerate kinase is necessary for activity and ...
|
|
14164
|
The fast-reacting sulfhydryl group of rat muscle phosphoglycerate kinase is necessary for activity and ...
|
|
14165
|
The fast-reacting sulfhydryl group of rat muscle phosphoglycerate kinase is necessary for activity and ...
|
|
14166
|
The fast-reacting sulfhydryl group of rat muscle phosphoglycerate kinase is necessary for activity and ...
|
|
14167
|
The fast-reacting sulfhydryl group of rat muscle phosphoglycerate kinase is necessary for activity and ...
|
|
14168
|
Construction of a synthetic gene for an R-plasmid-encoded dihydrofolate reductase and studies on the role of ...
|
|
14169
|
Construction of a synthetic gene for an R-plasmid-encoded dihydrofolate reductase and studies on the role of ...
|
|
14170
|
Construction of a synthetic gene for an R-plasmid-encoded dihydrofolate reductase and studies on the role of ...
|
|
14171
|
Construction of a synthetic gene for an R-plasmid-encoded dihydrofolate reductase and studies on the role of ...
|
|
14189
|
Kinetic Studies on the reaction catalyzed by Phosphoglycerate Kinase. I. The Effect of Mg2+ and Adenosine ...
|
|
14190
|
Kinetic Studies on the reaction catalyzed by Phosphoglycerate Kinase. I. The Effect of Mg2+ and Adenosine ...
|
|
14191
|
Kinetic Studies on the reaction catalyzed by Phosphoglycerate Kinase. I. The Effect of Mg2+ and Adenosine ...
|
|
14192
|
Kinetic Studies on the reaction catalyzed by Phosphoglycerate Kinase. I. The Effect of Mg2+ and Adenosine ...
|
|
14193
|
Kinetic Studies on the reaction catalyzed by Phosphoglycerate Kinase. I. The Effect of Mg2+ and Adenosine ...
|
|
14194
|
Kinetic Studies on the reaction catalyzed by Phosphoglycerate Kinase. I. The Effect of Mg2+ and Adenosine ...
|
|
14195
|
Kinetic Studies on the reaction catalyzed by Phosphoglycerate Kinase. I. The Effect of Mg2+ and Adenosine ...
|
|
14196
|
Kinetic Studies on the reaction catalyzed by Phosphoglycerate Kinase. I. The Effect of Mg2+ and Adenosine ...
|
|
14197
|
Kinetic Studies on the reaction catalyzed by Phosphoglycerate Kinase. I. The Effect of Mg2+ and Adenosine ...
|
|
14198
|
Kinetic Studies on the reaction catalyzed by Phosphoglycerate Kinase. I. The Effect of Mg2+ and Adenosine ...
|
|
14199
|
Kinetic Studies on the reaction catalyzed by Phosphoglycerate Kinase. I. The Effect of Mg2+ and Adenosine ...
|
|
14200
|
Kinetic Studies on the reaction catalyzed by Phosphoglycerate Kinase. I. The Effect of Mg2+ and Adenosine ...
|
|
14201
|
Kinetic Studies on the reaction catalyzed by Phosphoglycerate Kinase. I. The Effect of Mg2+ and Adenosine ...
|
|
14202
|
Kinetic Studies on the reaction catalyzed by Phosphoglycerate Kinase. I. The Effect of Mg2+ and Adenosine ...
|
|
14203
|
Kinetic Studies on the reaction catalyzed by Phosphoglycerate Kinase. I. The Effect of Mg2+ and Adenosine ...
|
|
14204
|
Properties of uridine diphosphate glucose pyrophosphorylase from Golgi apparatus of liver
|
|
14205
|
Properties of uridine diphosphate glucose pyrophosphorylase from Golgi apparatus of liver
|
|
14206
|
Properties of uridine diphosphate glucose pyrophosphorylase from Golgi apparatus of liver
|
|
14207
|
Properties of uridine diphosphate glucose pyrophosphorylase from Golgi apparatus of liver
|
|
14208
|
The contribution of lysine-36 to catalysis by human myo-inositol monophosphatase
|
|
14209
|
The contribution of lysine-36 to catalysis by human myo-inositol monophosphatase
|
|
14210
|
The contribution of lysine-36 to catalysis by human myo-inositol monophosphatase
|
|
14211
|
The contribution of lysine-36 to catalysis by human myo-inositol monophosphatase
|
|
14212
|
The contribution of lysine-36 to catalysis by human myo-inositol monophosphatase
|
|
14213
|
The contribution of lysine-36 to catalysis by human myo-inositol monophosphatase
|
|
14214
|
The contribution of lysine-36 to catalysis by human myo-inositol monophosphatase
|
|
14215
|
The contribution of lysine-36 to catalysis by human myo-inositol monophosphatase
|
|
14216
|
The contribution of lysine-36 to catalysis by human myo-inositol monophosphatase
|
|
14217
|
The contribution of lysine-36 to catalysis by human myo-inositol monophosphatase
|
|
14218
|
The contribution of lysine-36 to catalysis by human myo-inositol monophosphatase
|
|
14219
|
The contribution of lysine-36 to catalysis by human myo-inositol monophosphatase
|
|
14220
|
The contribution of lysine-36 to catalysis by human myo-inositol monophosphatase
|
|
14221
|
The contribution of lysine-36 to catalysis by human myo-inositol monophosphatase
|
|
14222
|
The contribution of lysine-36 to catalysis by human myo-inositol monophosphatase
|
|
14223
|
The contribution of lysine-36 to catalysis by human myo-inositol monophosphatase
|
|
14224
|
Electrostatic stabilization in a pre-organized polar active site: the catalytic role of Lys-80 in Candida ...
|
|
14225
|
Electrostatic stabilization in a pre-organized polar active site: the catalytic role of Lys-80 in Candida ...
|
|
14226
|
Electrostatic stabilization in a pre-organized polar active site: the catalytic role of Lys-80 in Candida ...
|
|
14227
|
Electrostatic stabilization in a pre-organized polar active site: the catalytic role of Lys-80 in Candida ...
|
|
14228
|
Electrostatic stabilization in a pre-organized polar active site: the catalytic role of Lys-80 in Candida ...
|
|
14229
|
Electrostatic stabilization in a pre-organized polar active site: the catalytic role of Lys-80 in Candida ...
|
|
14230
|
Electrostatic stabilization in a pre-organized polar active site: the catalytic role of Lys-80 in Candida ...
|
|
14231
|
Electrostatic stabilization in a pre-organized polar active site: the catalytic role of Lys-80 in Candida ...
|
|
14232
|
Electrostatic stabilization in a pre-organized polar active site: the catalytic role of Lys-80 in Candida ...
|
|
14233
|
Electrostatic stabilization in a pre-organized polar active site: the catalytic role of Lys-80 in Candida ...
|
|
14234
|
Electrostatic stabilization in a pre-organized polar active site: the catalytic role of Lys-80 in Candida ...
|
|
14235
|
Electrostatic stabilization in a pre-organized polar active site: the catalytic role of Lys-80 in Candida ...
|
|
14236
|
Electrostatic stabilization in a pre-organized polar active site: the catalytic role of Lys-80 in Candida ...
|
|
14237
|
Electrostatic stabilization in a pre-organized polar active site: the catalytic role of Lys-80 in Candida ...
|
|
14238
|
Electrostatic stabilization in a pre-organized polar active site: the catalytic role of Lys-80 in Candida ...
|
|
14239
|
Electrostatic stabilization in a pre-organized polar active site: the catalytic role of Lys-80 in Candida ...
|
|
14240
|
Electrostatic stabilization in a pre-organized polar active site: the catalytic role of Lys-80 in Candida ...
|
|
14241
|
Electrostatic stabilization in a pre-organized polar active site: the catalytic role of Lys-80 in Candida ...
|
|
14242
|
Electrostatic stabilization in a pre-organized polar active site: the catalytic role of Lys-80 in Candida ...
|
|
14243
|
Electrostatic stabilization in a pre-organized polar active site: the catalytic role of Lys-80 in Candida ...
|
|
14244
|
Electrostatic stabilization in a pre-organized polar active site: the catalytic role of Lys-80 in Candida ...
|
|
14245
|
Electrostatic stabilization in a pre-organized polar active site: the catalytic role of Lys-80 in Candida ...
|
|
14246
|
Phosphofructokinase from Lactobacteriaceae II. Purification and Properties of Phosphofructokinase from ...
|
|
14247
|
Phosphofructokinase from Lactobacteriaceae II. Purification and Properties of Phosphofructokinase from ...
|
|
14248
|
Phosphofructokinase from Lactobacteriaceae II. Purification and Properties of Phosphofructokinase from ...
|
|
14249
|
Phosphofructokinase from Lactobacteriaceae II. Purification and Properties of Phosphofructokinase from ...
|
|
14250
|
Phosphofructokinase from Lactobacteriaceae II. Purification and Properties of Phosphofructokinase from ...
|
|
14251
|
Phosphofructokinase from Lactobacteriaceae II. Purification and Properties of Phosphofructokinase from ...
|
|
14252
|
Catalytic properties of the archaeal S-adenosylmethionine decarboxylase from Methanococcus jannaschii
|
|
14253
|
Catalytic properties of the archaeal S-adenosylmethionine decarboxylase from Methanococcus jannaschii
|
|
14254
|
Catalytic properties of the archaeal S-adenosylmethionine decarboxylase from Methanococcus jannaschii
|
|
14255
|
Isolation and biochemical characterization of two soluble iron(III) reductases from Paracoccus denitrificans
|
|
14256
|
Isolation and biochemical characterization of two soluble iron(III) reductases from Paracoccus denitrificans
|
|
14257
|
Isolation and biochemical characterization of two soluble iron(III) reductases from Paracoccus denitrificans
|
|
14258
|
Isolation and biochemical characterization of two soluble iron(III) reductases from Paracoccus denitrificans
|
|
14259
|
Isolation and biochemical characterization of two soluble iron(III) reductases from Paracoccus denitrificans
|
|
14260
|
Isolation and biochemical characterization of two soluble iron(III) reductases from Paracoccus denitrificans
|
|
14261
|
Isolation and biochemical characterization of two soluble iron(III) reductases from Paracoccus denitrificans
|
|
14262
|
Isolation and biochemical characterization of two soluble iron(III) reductases from Paracoccus denitrificans
|
|
14263
|
Isolation and biochemical characterization of two soluble iron(III) reductases from Paracoccus denitrificans
|
|
14264
|
Isolation and biochemical characterization of two soluble iron(III) reductases from Paracoccus denitrificans
|
|
14265
|
Purification and properties of fructokinase I from Lactococcus lactis. Localization of scrK on the ...
|
|
14266
|
Purification and properties of fructokinase I from Lactococcus lactis. Localization of scrK on the ...
|
|
14267
|
Purification and properties of fructokinase I from Lactococcus lactis. Localization of scrK on the ...
|
|
14268
|
Characterization of aromatic aminotransferases from the hyperthermophilic archaeon Thermococcus litoralis
|
|
14269
|
Characterization of aromatic aminotransferases from the hyperthermophilic archaeon Thermococcus litoralis
|
|
14270
|
Characterization of aromatic aminotransferases from the hyperthermophilic archaeon Thermococcus litoralis
|
|
14271
|
Characterization of aromatic aminotransferases from the hyperthermophilic archaeon Thermococcus litoralis
|
|
14272
|
Characterization of aromatic aminotransferases from the hyperthermophilic archaeon Thermococcus litoralis
|
|
14273
|
Characterization of aromatic aminotransferases from the hyperthermophilic archaeon Thermococcus litoralis
|
|
14274
|
Structure and mechanism of glutamate racemase from Aquifex pyrophilus
|
|
14275
|
Structure and mechanism of glutamate racemase from Aquifex pyrophilus
|
|
14276
|
Structure and mechanism of glutamate racemase from Aquifex pyrophilus
|
|
14277
|
Structure and mechanism of glutamate racemase from Aquifex pyrophilus
|
|
14278
|
Structure and mechanism of glutamate racemase from Aquifex pyrophilus
|
|
14279
|
Structure and mechanism of glutamate racemase from Aquifex pyrophilus
|
|
14280
|
A transient intermediate in the reaction catalyzed by (S)-mandelate dehydrogenase from Pseudomonas putida
|
|
14281
|
A transient intermediate in the reaction catalyzed by (S)-mandelate dehydrogenase from Pseudomonas putida
|
|
14282
|
Enzymatic proof for the identity of the S-sulfocysteine synthase and cysteine synthase B of Salmonella ...
|
|
14283
|
Enzymatic proof for the identity of the S-sulfocysteine synthase and cysteine synthase B of Salmonella ...
|
|
14284
|
Single-turnover kinetics of homoprotocatechuate 2,3-dioxygenase
|
|
14285
|
Single-turnover kinetics of homoprotocatechuate 2,3-dioxygenase
|
|
14286
|
Single-turnover kinetics of homoprotocatechuate 2,3-dioxygenase
|
|
14287
|
Single-turnover kinetics of homoprotocatechuate 2,3-dioxygenase
|
|
14288
|
Magnesium ion requirements for yeast enolase activity
|
|
14289
|
Magnesium ion requirements for yeast enolase activity
|
|
14290
|
Mutational analysis of the catalytic domain of O-linked N-acetylglucosaminyl transferase
|
|
14291
|
Mutational analysis of the catalytic domain of O-linked N-acetylglucosaminyl transferase
|
|
14292
|
Mutational analysis of the catalytic domain of O-linked N-acetylglucosaminyl transferase
|
|
14293
|
Mutational analysis of the catalytic domain of O-linked N-acetylglucosaminyl transferase
|
|
14294
|
Mutational analysis of the catalytic domain of O-linked N-acetylglucosaminyl transferase
|
|
14295
|
Tyrosine residues near the FAD binding site are critical for FAD binding and for the maintenance of the stable ...
|
|
14296
|
Tyrosine residues near the FAD binding site are critical for FAD binding and for the maintenance of the stable ...
|
|
14297
|
Tyrosine residues near the FAD binding site are critical for FAD binding and for the maintenance of the stable ...
|
|
14298
|
Tyrosine residues near the FAD binding site are critical for FAD binding and for the maintenance of the stable ...
|
|
14299
|
Tyrosine residues near the FAD binding site are critical for FAD binding and for the maintenance of the stable ...
|
|
14300
|
Tyrosine residues near the FAD binding site are critical for FAD binding and for the maintenance of the stable ...
|
|
14301
|
Tyrosine residues near the FAD binding site are critical for FAD binding and for the maintenance of the stable ...
|
|
14302
|
Tyrosine residues near the FAD binding site are critical for FAD binding and for the maintenance of the stable ...
|
|
14303
|
Tyrosine residues near the FAD binding site are critical for FAD binding and for the maintenance of the stable ...
|
|
14304
|
Tyrosine residues near the FAD binding site are critical for FAD binding and for the maintenance of the stable ...
|
|
14305
|
Tyrosine residues near the FAD binding site are critical for FAD binding and for the maintenance of the stable ...
|
|
14306
|
Tyrosine residues near the FAD binding site are critical for FAD binding and for the maintenance of the stable ...
|
|
14307
|
Tyrosine residues near the FAD binding site are critical for FAD binding and for the maintenance of the stable ...
|
|
14308
|
Tyrosine residues near the FAD binding site are critical for FAD binding and for the maintenance of the stable ...
|
|
14309
|
Tyrosine residues near the FAD binding site are critical for FAD binding and for the maintenance of the stable ...
|
|
14310
|
Tyrosine residues near the FAD binding site are critical for FAD binding and for the maintenance of the stable ...
|
|
14311
|
Tyrosine residues near the FAD binding site are critical for FAD binding and for the maintenance of the stable ...
|
|
14312
|
Tyrosine residues near the FAD binding site are critical for FAD binding and for the maintenance of the stable ...
|
|
14313
|
Tyrosine residues near the FAD binding site are critical for FAD binding and for the maintenance of the stable ...
|
|
14314
|
Tyrosine residues near the FAD binding site are critical for FAD binding and for the maintenance of the stable ...
|
|
14315
|
Tyrosine residues near the FAD binding site are critical for FAD binding and for the maintenance of the stable ...
|
|
14316
|
Site-directed mutagenesis of catalytic residues of influenza virus neuraminidase as an aid to drug design
|
|
14317
|
Site-directed mutagenesis of catalytic residues of influenza virus neuraminidase as an aid to drug design
|
|
14318
|
Site-directed mutagenesis of catalytic residues of influenza virus neuraminidase as an aid to drug design
|
|
14319
|
Site-directed mutagenesis of catalytic residues of influenza virus neuraminidase as an aid to drug design
|
|
14320
|
Site-directed mutagenesis of catalytic residues of influenza virus neuraminidase as an aid to drug design
|
|
14321
|
Site-directed mutagenesis of catalytic residues of influenza virus neuraminidase as an aid to drug design
|
|
14322
|
Site-directed mutagenesis of catalytic residues of influenza virus neuraminidase as an aid to drug design
|
|
14323
|
Site-directed mutagenesis of catalytic residues of influenza virus neuraminidase as an aid to drug design
|
|
14324
|
Site-directed mutagenesis of catalytic residues of influenza virus neuraminidase as an aid to drug design
|
|
14325
|
Site-directed mutagenesis of catalytic residues of influenza virus neuraminidase as an aid to drug design
|
|
14326
|
Site-directed mutagenesis of catalytic residues of influenza virus neuraminidase as an aid to drug design
|
|
14327
|
Site-directed mutagenesis of catalytic residues of influenza virus neuraminidase as an aid to drug design
|
|
14328
|
Site-directed mutagenesis of catalytic residues of influenza virus neuraminidase as an aid to drug design
|
|
14329
|
Site-directed mutagenesis of catalytic residues of influenza virus neuraminidase as an aid to drug design
|
|
14330
|
Site-directed mutagenesis of catalytic residues of influenza virus neuraminidase as an aid to drug design
|
|
14331
|
Site-directed mutagenesis of catalytic residues of influenza virus neuraminidase as an aid to drug design
|
|
14332
|
Site-directed mutagenesis of catalytic residues of influenza virus neuraminidase as an aid to drug design
|
|
14333
|
Catalytic and structural characteristics of carp hepatopancreas D-amino acid oxidase expressed in Escherichia ...
|
|
14334
|
Catalytic and structural characteristics of carp hepatopancreas D-amino acid oxidase expressed in Escherichia ...
|
|
14335
|
Catalytic and structural characteristics of carp hepatopancreas D-amino acid oxidase expressed in Escherichia ...
|
|
14336
|
Catalytic and structural characteristics of carp hepatopancreas D-amino acid oxidase expressed in Escherichia ...
|
|
14337
|
Catalytic and structural characteristics of carp hepatopancreas D-amino acid oxidase expressed in Escherichia ...
|
|
14338
|
Catalytic and structural characteristics of carp hepatopancreas D-amino acid oxidase expressed in Escherichia ...
|
|
14339
|
Catalytic and structural characteristics of carp hepatopancreas D-amino acid oxidase expressed in Escherichia ...
|
|
14340
|
Catalytic and structural characteristics of carp hepatopancreas D-amino acid oxidase expressed in Escherichia ...
|
|
14341
|
Catalytic and structural characteristics of carp hepatopancreas D-amino acid oxidase expressed in Escherichia ...
|
|
14342
|
Catalytic and structural characteristics of carp hepatopancreas D-amino acid oxidase expressed in Escherichia ...
|
|
14343
|
Catalytic and structural characteristics of carp hepatopancreas D-amino acid oxidase expressed in Escherichia ...
|
|
14344
|
Catalytic and structural characteristics of carp hepatopancreas D-amino acid oxidase expressed in Escherichia ...
|
|
14345
|
Characterization of recombinant diaminopropionate ammonia-lyase from Escherichia coli and Salmonella ...
|
|
14346
|
Characterization of recombinant diaminopropionate ammonia-lyase from Escherichia coli and Salmonella ...
|
|
14347
|
Characterization of recombinant diaminopropionate ammonia-lyase from Escherichia coli and Salmonella ...
|
|
14348
|
Characterization of recombinant diaminopropionate ammonia-lyase from Escherichia coli and Salmonella ...
|
|
14349
|
Characterization of recombinant diaminopropionate ammonia-lyase from Escherichia coli and Salmonella ...
|
|
14350
|
Characterization of recombinant diaminopropionate ammonia-lyase from Escherichia coli and Salmonella ...
|
|
14351
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14352
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14353
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14354
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14355
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14356
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14357
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14358
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14359
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14360
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14361
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14362
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14363
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14364
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14365
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14366
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14367
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14368
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14369
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14370
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14371
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14372
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14373
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14374
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14375
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14376
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14377
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14378
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14379
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14380
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14381
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14382
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14383
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14384
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14385
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14386
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14387
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14388
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14389
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14390
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14391
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14392
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14393
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14394
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14395
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14396
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14397
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14398
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14399
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14400
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14401
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14402
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14403
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14404
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14405
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14406
|
The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication
|
|
14407
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14408
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14409
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14410
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14411
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14412
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14413
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14414
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14415
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14416
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14417
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14418
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14419
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14420
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14421
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14422
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14423
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14424
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14425
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14426
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14427
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14428
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14429
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14430
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14431
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14432
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14433
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14434
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14435
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14436
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14437
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14438
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14439
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14440
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14441
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14442
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14443
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14444
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14445
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14446
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14447
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14448
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14449
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14450
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14451
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14452
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14453
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14454
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14455
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14456
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14457
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14458
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14459
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14460
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14461
|
Ornithine transcarbamylase from Streptococcus faecalis and bovine liver. II. Multiple binding sites for ...
|
|
14462
|
Interaction of D-fructose and fructose 1-phosphate with yeast phosphofructokinase and its influence on ...
|
|
14463
|
Interaction of D-fructose and fructose 1-phosphate with yeast phosphofructokinase and its influence on ...
|
|
14464
|
Interaction of D-fructose and fructose 1-phosphate with yeast phosphofructokinase and its influence on ...
|
|
14465
|
Interaction of D-fructose and fructose 1-phosphate with yeast phosphofructokinase and its influence on ...
|
|
14466
|
Interaction of D-fructose and fructose 1-phosphate with yeast phosphofructokinase and its influence on ...
|
|
14467
|
Interaction of D-fructose and fructose 1-phosphate with yeast phosphofructokinase and its influence on ...
|
|
14468
|
Interaction of D-fructose and fructose 1-phosphate with yeast phosphofructokinase and its influence on ...
|
|
14469
|
Interaction of D-fructose and fructose 1-phosphate with yeast phosphofructokinase and its influence on ...
|
|
14470
|
Interaction of D-fructose and fructose 1-phosphate with yeast phosphofructokinase and its influence on ...
|
|
14471
|
Interaction of D-fructose and fructose 1-phosphate with yeast phosphofructokinase and its influence on ...
|
|
14472
|
Functional characterization of Delta3,Delta2-enoyl-CoA isomerases from rat liver
|
|
14473
|
Functional characterization of Delta3,Delta2-enoyl-CoA isomerases from rat liver
|
|
14474
|
Functional characterization of Delta3,Delta2-enoyl-CoA isomerases from rat liver
|
|
14475
|
Functional characterization of Delta3,Delta2-enoyl-CoA isomerases from rat liver
|
|
14476
|
Functional characterization of Delta3,Delta2-enoyl-CoA isomerases from rat liver
|
|
14477
|
Functional characterization of Delta3,Delta2-enoyl-CoA isomerases from rat liver
|
|
14478
|
Functional characterization of Delta3,Delta2-enoyl-CoA isomerases from rat liver
|
|
14479
|
Functional characterization of Delta3,Delta2-enoyl-CoA isomerases from rat liver
|
|
14480
|
Functional characterization of Delta3,Delta2-enoyl-CoA isomerases from rat liver
|
|
14481
|
Functional characterization of Delta3,Delta2-enoyl-CoA isomerases from rat liver
|
|
14482
|
Functional characterization of Delta3,Delta2-enoyl-CoA isomerases from rat liver
|
|
14483
|
Functional characterization of Delta3,Delta2-enoyl-CoA isomerases from rat liver
|
|
14484
|
Functional characterization of Delta3,Delta2-enoyl-CoA isomerases from rat liver
|
|
14485
|
Functional characterization of Delta3,Delta2-enoyl-CoA isomerases from rat liver
|
|
14486
|
Functional characterization of Delta3,Delta2-enoyl-CoA isomerases from rat liver
|
|
14487
|
Functional characterization of Delta3,Delta2-enoyl-CoA isomerases from rat liver
|
|
14488
|
Functional characterization of Delta3,Delta2-enoyl-CoA isomerases from rat liver
|
|
14489
|
Functional characterization of Delta3,Delta2-enoyl-CoA isomerases from rat liver
|
|
14490
|
Functional characterization of Delta3,Delta2-enoyl-CoA isomerases from rat liver
|
|
14491
|
Functional characterization of Delta3,Delta2-enoyl-CoA isomerases from rat liver
|
|
14492
|
Functional characterization of Delta3,Delta2-enoyl-CoA isomerases from rat liver
|
|
14493
|
Functional characterization of Delta3,Delta2-enoyl-CoA isomerases from rat liver
|
|
14494
|
Functional characterization of Delta3,Delta2-enoyl-CoA isomerases from rat liver
|
|
14495
|
Functional characterization of Delta3,Delta2-enoyl-CoA isomerases from rat liver
|
|
14496
|
Human platelet 6-phosphofructokinase. Relation between inhibition by Mg-ATP2-and cooperativity towards ...
|
|
14497
|
Human platelet 6-phosphofructokinase. Relation between inhibition by Mg-ATP2-and cooperativity towards ...
|
|
14498
|
Human platelet 6-phosphofructokinase. Relation between inhibition by Mg-ATP2-and cooperativity towards ...
|
|
14499
|
Human platelet 6-phosphofructokinase. Relation between inhibition by Mg-ATP2-and cooperativity towards ...
|
|
14500
|
Human platelet 6-phosphofructokinase. Relation between inhibition by Mg-ATP2-and cooperativity towards ...
|
|
14501
|
Human platelet 6-phosphofructokinase. Relation between inhibition by Mg-ATP2-and cooperativity towards ...
|
|
14502
|
Human platelet 6-phosphofructokinase. Relation between inhibition by Mg-ATP2-and cooperativity towards ...
|
|
14503
|
Human platelet 6-phosphofructokinase. Relation between inhibition by Mg-ATP2-and cooperativity towards ...
|
|
14504
|
Human platelet 6-phosphofructokinase. Relation between inhibition by Mg-ATP2-and cooperativity towards ...
|
|
14505
|
Human platelet 6-phosphofructokinase. Relation between inhibition by Mg-ATP2-and cooperativity towards ...
|
|
14506
|
A purification of microsomal glucose-6-phosphatase from human tissue
|
|
14507
|
A purification of microsomal glucose-6-phosphatase from human tissue
|
|
14508
|
Cytosolic and mitochondrial deoxyribonucleotidases: activity with substrate analogs, inhibitors and ...
|
|
14509
|
Cytosolic and mitochondrial deoxyribonucleotidases: activity with substrate analogs, inhibitors and ...
|
|
14510
|
Cytosolic and mitochondrial deoxyribonucleotidases: activity with substrate analogs, inhibitors and ...
|
|
14511
|
Cytosolic and mitochondrial deoxyribonucleotidases: activity with substrate analogs, inhibitors and ...
|
|
14512
|
Cytosolic and mitochondrial deoxyribonucleotidases: activity with substrate analogs, inhibitors and ...
|
|
14513
|
Cytosolic and mitochondrial deoxyribonucleotidases: activity with substrate analogs, inhibitors and ...
|
|
14514
|
Cytosolic and mitochondrial deoxyribonucleotidases: activity with substrate analogs, inhibitors and ...
|
|
14515
|
Cytosolic and mitochondrial deoxyribonucleotidases: activity with substrate analogs, inhibitors and ...
|
|
14516
|
Cytosolic and mitochondrial deoxyribonucleotidases: activity with substrate analogs, inhibitors and ...
|
|
14517
|
Cytosolic and mitochondrial deoxyribonucleotidases: activity with substrate analogs, inhibitors and ...
|
|
14518
|
Cytosolic and mitochondrial deoxyribonucleotidases: activity with substrate analogs, inhibitors and ...
|
|
14519
|
Cytosolic and mitochondrial deoxyribonucleotidases: activity with substrate analogs, inhibitors and ...
|
|
14520
|
Cytosolic and mitochondrial deoxyribonucleotidases: activity with substrate analogs, inhibitors and ...
|
|
14521
|
Cytosolic and mitochondrial deoxyribonucleotidases: activity with substrate analogs, inhibitors and ...
|
|
14522
|
Cytosolic and mitochondrial deoxyribonucleotidases: activity with substrate analogs, inhibitors and ...
|
|
14523
|
Cytosolic and mitochondrial deoxyribonucleotidases: activity with substrate analogs, inhibitors and ...
|
|
14524
|
Cytosolic and mitochondrial deoxyribonucleotidases: activity with substrate analogs, inhibitors and ...
|
|
14525
|
Cytosolic and mitochondrial deoxyribonucleotidases: activity with substrate analogs, inhibitors and ...
|
|
14526
|
Cytosolic and mitochondrial deoxyribonucleotidases: activity with substrate analogs, inhibitors and ...
|
|
14527
|
Reactivity and metal-dependent stereospecificity of the phosphorothioate analogs of ADP and ATP and reactivity ...
|
|
14528
|
Reactivity and metal-dependent stereospecificity of the phosphorothioate analogs of ADP and ATP and reactivity ...
|
|
14529
|
Reactivity and metal-dependent stereospecificity of the phosphorothioate analogs of ADP and ATP and reactivity ...
|
|
14530
|
Reactivity and metal-dependent stereospecificity of the phosphorothioate analogs of ADP and ATP and reactivity ...
|
|
14531
|
Reactivity and metal-dependent stereospecificity of the phosphorothioate analogs of ADP and ATP and reactivity ...
|
|
14532
|
Reactivity and metal-dependent stereospecificity of the phosphorothioate analogs of ADP and ATP and reactivity ...
|
|
14533
|
Reactivity and metal-dependent stereospecificity of the phosphorothioate analogs of ADP and ATP and reactivity ...
|
|
14534
|
Reactivity and metal-dependent stereospecificity of the phosphorothioate analogs of ADP and ATP and reactivity ...
|
|
14535
|
Reactivity and metal-dependent stereospecificity of the phosphorothioate analogs of ADP and ATP and reactivity ...
|
|
14536
|
Reactivity and metal-dependent stereospecificity of the phosphorothioate analogs of ADP and ATP and reactivity ...
|
|
14537
|
Reactivity and metal-dependent stereospecificity of the phosphorothioate analogs of ADP and ATP and reactivity ...
|
|
14538
|
Reactivity and metal-dependent stereospecificity of the phosphorothioate analogs of ADP and ATP and reactivity ...
|
|
14539
|
Reactivity and metal-dependent stereospecificity of the phosphorothioate analogs of ADP and ATP and reactivity ...
|
|
14540
|
Reactivity and metal-dependent stereospecificity of the phosphorothioate analogs of ADP and ATP and reactivity ...
|
|
14541
|
Reactivity and metal-dependent stereospecificity of the phosphorothioate analogs of ADP and ATP and reactivity ...
|
|
14542
|
Reactivity and metal-dependent stereospecificity of the phosphorothioate analogs of ADP and ATP and reactivity ...
|
|
14543
|
Reactivity and metal-dependent stereospecificity of the phosphorothioate analogs of ADP and ATP and reactivity ...
|
|
14544
|
Comparison of the stability and substrate specificity of purified peroxisomal 3-oxoacyl-CoA thiolases A and B ...
|
|
14545
|
Comparison of the stability and substrate specificity of purified peroxisomal 3-oxoacyl-CoA thiolases A and B ...
|
|
14546
|
Comparison of the stability and substrate specificity of purified peroxisomal 3-oxoacyl-CoA thiolases A and B ...
|
|
14547
|
Comparison of the stability and substrate specificity of purified peroxisomal 3-oxoacyl-CoA thiolases A and B ...
|
|
14548
|
Comparison of the stability and substrate specificity of purified peroxisomal 3-oxoacyl-CoA thiolases A and B ...
|
|
14549
|
Comparison of the stability and substrate specificity of purified peroxisomal 3-oxoacyl-CoA thiolases A and B ...
|
|
14550
|
Comparison of the stability and substrate specificity of purified peroxisomal 3-oxoacyl-CoA thiolases A and B ...
|
|
14551
|
Comparison of the stability and substrate specificity of purified peroxisomal 3-oxoacyl-CoA thiolases A and B ...
|
|
14552
|
Mutational study on alphaGln90 of Fe-type nitrile hydratase from Rhodococcus sp. N771
|
|
14553
|
Mutational study on alphaGln90 of Fe-type nitrile hydratase from Rhodococcus sp. N771
|
|
14554
|
Mutational study on alphaGln90 of Fe-type nitrile hydratase from Rhodococcus sp. N771
|
|
14555
|
Mutational study on alphaGln90 of Fe-type nitrile hydratase from Rhodococcus sp. N771
|
|
14556
|
Mutational study on alphaGln90 of Fe-type nitrile hydratase from Rhodococcus sp. N771
|
|
14557
|
Mutational study on alphaGln90 of Fe-type nitrile hydratase from Rhodococcus sp. N771
|
|
14558
|
Mutational study on alphaGln90 of Fe-type nitrile hydratase from Rhodococcus sp. N771
|
|
14559
|
Mutational study on alphaGln90 of Fe-type nitrile hydratase from Rhodococcus sp. N771
|
|
14560
|
Substrate specificities of 3-oxoacyl-CoA thiolase A and sterol carrier protein 2/3-oxoacyl-CoA thiolase ...
|
|
14561
|
Substrate specificities of 3-oxoacyl-CoA thiolase A and sterol carrier protein 2/3-oxoacyl-CoA thiolase ...
|
|
14562
|
Substrate specificities of 3-oxoacyl-CoA thiolase A and sterol carrier protein 2/3-oxoacyl-CoA thiolase ...
|
|
14563
|
Substrate specificities of 3-oxoacyl-CoA thiolase A and sterol carrier protein 2/3-oxoacyl-CoA thiolase ...
|
|
14564
|
Substrate specificities of 3-oxoacyl-CoA thiolase A and sterol carrier protein 2/3-oxoacyl-CoA thiolase ...
|
|
14565
|
Substrate specificities of 3-oxoacyl-CoA thiolase A and sterol carrier protein 2/3-oxoacyl-CoA thiolase ...
|
|
14566
|
Substrate specificities of 3-oxoacyl-CoA thiolase A and sterol carrier protein 2/3-oxoacyl-CoA thiolase ...
|
|
14567
|
Substrate specificities of 3-oxoacyl-CoA thiolase A and sterol carrier protein 2/3-oxoacyl-CoA thiolase ...
|
|
14568
|
Substrate specificities of 3-oxoacyl-CoA thiolase A and sterol carrier protein 2/3-oxoacyl-CoA thiolase ...
|
|
14569
|
Substrate specificities of 3-oxoacyl-CoA thiolase A and sterol carrier protein 2/3-oxoacyl-CoA thiolase ...
|
|
14570
|
Substrate specificities of 3-oxoacyl-CoA thiolase A and sterol carrier protein 2/3-oxoacyl-CoA thiolase ...
|
|
14571
|
Substrate specificities of 3-oxoacyl-CoA thiolase A and sterol carrier protein 2/3-oxoacyl-CoA thiolase ...
|
|
14572
|
Substrate specificities of 3-oxoacyl-CoA thiolase A and sterol carrier protein 2/3-oxoacyl-CoA thiolase ...
|
|
14573
|
Substrate specificities of 3-oxoacyl-CoA thiolase A and sterol carrier protein 2/3-oxoacyl-CoA thiolase ...
|
|
14574
|
Substrate specificities of 3-oxoacyl-CoA thiolase A and sterol carrier protein 2/3-oxoacyl-CoA thiolase ...
|
|
14575
|
Substrate specificities of 3-oxoacyl-CoA thiolase A and sterol carrier protein 2/3-oxoacyl-CoA thiolase ...
|
|
14576
|
Substrate specificities of 3-oxoacyl-CoA thiolase A and sterol carrier protein 2/3-oxoacyl-CoA thiolase ...
|
|
14577
|
Substrate specificities of 3-oxoacyl-CoA thiolase A and sterol carrier protein 2/3-oxoacyl-CoA thiolase ...
|
|
14578
|
Substrate specificities of 3-oxoacyl-CoA thiolase A and sterol carrier protein 2/3-oxoacyl-CoA thiolase ...
|
|
14579
|
Characterization of human UMP/CMP kinase and its phosphorylation of D- and L-form deoxycytidine analogue ...
|
|
14580
|
Characterization of human UMP/CMP kinase and its phosphorylation of D- and L-form deoxycytidine analogue ...
|
|
14581
|
Characterization of human UMP/CMP kinase and its phosphorylation of D- and L-form deoxycytidine analogue ...
|
|
14582
|
Characterization of human UMP/CMP kinase and its phosphorylation of D- and L-form deoxycytidine analogue ...
|
|
14583
|
Characterization of human UMP/CMP kinase and its phosphorylation of D- and L-form deoxycytidine analogue ...
|
|
14584
|
Characterization of human UMP/CMP kinase and its phosphorylation of D- and L-form deoxycytidine analogue ...
|
|
14585
|
Characterization of human UMP/CMP kinase and its phosphorylation of D- and L-form deoxycytidine analogue ...
|
|
14586
|
Characterization of human UMP/CMP kinase and its phosphorylation of D- and L-form deoxycytidine analogue ...
|
|
14587
|
Characterization of human UMP/CMP kinase and its phosphorylation of D- and L-form deoxycytidine analogue ...
|
|
14588
|
Characterization of human UMP/CMP kinase and its phosphorylation of D- and L-form deoxycytidine analogue ...
|
|
14589
|
Characterization of human UMP/CMP kinase and its phosphorylation of D- and L-form deoxycytidine analogue ...
|
|
14590
|
Characterization of human UMP/CMP kinase and its phosphorylation of D- and L-form deoxycytidine analogue ...
|
|
14591
|
Characterization of human UMP/CMP kinase and its phosphorylation of D- and L-form deoxycytidine analogue ...
|
|
14592
|
Characterization of human UMP/CMP kinase and its phosphorylation of D- and L-form deoxycytidine analogue ...
|
|
14593
|
Characterization of human UMP/CMP kinase and its phosphorylation of D- and L-form deoxycytidine analogue ...
|
|
14594
|
Characterization of human UMP/CMP kinase and its phosphorylation of D- and L-form deoxycytidine analogue ...
|
|
14595
|
Characterization of human UMP/CMP kinase and its phosphorylation of D- and L-form deoxycytidine analogue ...
|
|
14596
|
Characterization of human UMP/CMP kinase and its phosphorylation of D- and L-form deoxycytidine analogue ...
|
|
14597
|
Properties of an affinity-column-purified human deoxycytidylate deaminase
|
|
14598
|
Properties of an affinity-column-purified human deoxycytidylate deaminase
|
|
14599
|
Properties of an affinity-column-purified human deoxycytidylate deaminase
|
|
14600
|
Properties of an affinity-column-purified human deoxycytidylate deaminase
|
|
14601
|
Rat dihydroorotate dehydrogenase: isolation of the recombinant enzyme from mitochondria of insect cells
|
|
14602
|
Rat dihydroorotate dehydrogenase: isolation of the recombinant enzyme from mitochondria of insect cells
|
|
14603
|
Rat dihydroorotate dehydrogenase: isolation of the recombinant enzyme from mitochondria of insect cells
|
|
14604
|
Rat dihydroorotate dehydrogenase: isolation of the recombinant enzyme from mitochondria of insect cells
|
|
14605
|
Rat dihydroorotate dehydrogenase: isolation of the recombinant enzyme from mitochondria of insect cells
|
|
14606
|
Rat dihydroorotate dehydrogenase: isolation of the recombinant enzyme from mitochondria of insect cells
|
|
14607
|
Rat dihydroorotate dehydrogenase: isolation of the recombinant enzyme from mitochondria of insect cells
|
|
14608
|
Rat dihydroorotate dehydrogenase: isolation of the recombinant enzyme from mitochondria of insect cells
|
|
14609
|
Identification and characterisation of human aldose 1-epimerase
|
|
14610
|
Identification and characterisation of human aldose 1-epimerase
|
|
14611
|
Identification and characterisation of human aldose 1-epimerase
|
|
14612
|
Identification and characterisation of human aldose 1-epimerase
|
|
14613
|
Identification and characterisation of human aldose 1-epimerase
|
|
14614
|
Identification and characterisation of human aldose 1-epimerase
|
|
14615
|
Identification and characterisation of human aldose 1-epimerase
|
|
14637
|
Assay method for Escherichia coli photolyase activity using single-strand cis-syn cyclobutane pyrimidine dimer ...
|
|
14638
|
Engineering the active center of the 6-phospho-beta-galactosidase from Lactococcus lactis
|
|
14639
|
Engineering the active center of the 6-phospho-beta-galactosidase from Lactococcus lactis
|
|
14640
|
Engineering the active center of the 6-phospho-beta-galactosidase from Lactococcus lactis
|
|
14641
|
Engineering the active center of the 6-phospho-beta-galactosidase from Lactococcus lactis
|
|
14642
|
Engineering the active center of the 6-phospho-beta-galactosidase from Lactococcus lactis
|
|
14643
|
Engineering the active center of the 6-phospho-beta-galactosidase from Lactococcus lactis
|
|
14644
|
Engineering the active center of the 6-phospho-beta-galactosidase from Lactococcus lactis
|
|
14645
|
Engineering the active center of the 6-phospho-beta-galactosidase from Lactococcus lactis
|
|
14646
|
Engineering the active center of the 6-phospho-beta-galactosidase from Lactococcus lactis
|
|
14647
|
Engineering the active center of the 6-phospho-beta-galactosidase from Lactococcus lactis
|
|
14648
|
Engineering the active center of the 6-phospho-beta-galactosidase from Lactococcus lactis
|
|
14649
|
Cloning and characterization of dipeptidyl peptidase 10, a new member of an emerging subgroup of serine ...
|
|
14650
|
Cloning and characterization of dipeptidyl peptidase 10, a new member of an emerging subgroup of serine ...
|
|
14662
|
Activation of Thermus phosphofructokinase by monovalent cations
|
|
14663
|
Activation of Thermus phosphofructokinase by monovalent cations
|
|
14664
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14665
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14666
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14667
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14668
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14669
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14670
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14671
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14672
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14673
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14674
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14675
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14676
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14677
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14678
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14679
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14680
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14681
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14682
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14683
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14684
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14685
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14686
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14687
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14688
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14689
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14690
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14691
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14692
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14693
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14694
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14695
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14696
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14697
|
Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate ...
|
|
14698
|
Human hereditary glutathione synthetase deficiency: kinetic properties of mutant enzymes
|
|
14699
|
Human hereditary glutathione synthetase deficiency: kinetic properties of mutant enzymes
|
|
14700
|
Human hereditary glutathione synthetase deficiency: kinetic properties of mutant enzymes
|
|
14701
|
Human hereditary glutathione synthetase deficiency: kinetic properties of mutant enzymes
|
|
14702
|
Human hereditary glutathione synthetase deficiency: kinetic properties of mutant enzymes
|
|
14703
|
Human hereditary glutathione synthetase deficiency: kinetic properties of mutant enzymes
|
|
14704
|
Human hereditary glutathione synthetase deficiency: kinetic properties of mutant enzymes
|
|
14705
|
Human hereditary glutathione synthetase deficiency: kinetic properties of mutant enzymes
|
|
14706
|
Human hereditary glutathione synthetase deficiency: kinetic properties of mutant enzymes
|
|
14707
|
Human hereditary glutathione synthetase deficiency: kinetic properties of mutant enzymes
|
|
14708
|
Human hereditary glutathione synthetase deficiency: kinetic properties of mutant enzymes
|
|
14709
|
Human hereditary glutathione synthetase deficiency: kinetic properties of mutant enzymes
|
|
14710
|
Human hereditary glutathione synthetase deficiency: kinetic properties of mutant enzymes
|
|
14711
|
Human hereditary glutathione synthetase deficiency: kinetic properties of mutant enzymes
|
|
14712
|
Human hereditary glutathione synthetase deficiency: kinetic properties of mutant enzymes
|
|
14713
|
Human hereditary glutathione synthetase deficiency: kinetic properties of mutant enzymes
|
|
14714
|
Human hereditary glutathione synthetase deficiency: kinetic properties of mutant enzymes
|
|
14715
|
Human hereditary glutathione synthetase deficiency: kinetic properties of mutant enzymes
|
|
14716
|
Human hereditary glutathione synthetase deficiency: kinetic properties of mutant enzymes
|
|
14717
|
Human hereditary glutathione synthetase deficiency: kinetic properties of mutant enzymes
|
|
14718
|
Human hereditary glutathione synthetase deficiency: kinetic properties of mutant enzymes
|
|
14719
|
Human hereditary glutathione synthetase deficiency: kinetic properties of mutant enzymes
|
|
14720
|
Human hereditary glutathione synthetase deficiency: kinetic properties of mutant enzymes
|
|
14721
|
Human hereditary glutathione synthetase deficiency: kinetic properties of mutant enzymes
|
|
14724
|
A defined subset of adenylyl cyclases is regulated by bicarbonate ion
|
|
14725
|
A defined subset of adenylyl cyclases is regulated by bicarbonate ion
|
|
14726
|
Broad substrate stereospecificity of the Mycobacterium tuberculosis 7-keto-8-aminopelargonic acid synthase: ...
|
|
14727
|
Broad substrate stereospecificity of the Mycobacterium tuberculosis 7-keto-8-aminopelargonic acid synthase: ...
|
|
14728
|
Broad substrate stereospecificity of the Mycobacterium tuberculosis 7-keto-8-aminopelargonic acid synthase: ...
|
|
14729
|
Broad substrate stereospecificity of the Mycobacterium tuberculosis 7-keto-8-aminopelargonic acid synthase: ...
|
|
14730
|
Broad substrate stereospecificity of the Mycobacterium tuberculosis 7-keto-8-aminopelargonic acid synthase: ...
|
|
14731
|
Nucleotide specificity of pyruvate kinase and phosphoenolpyruvate carboxykinase
|
|
14732
|
Nucleotide specificity of pyruvate kinase and phosphoenolpyruvate carboxykinase
|
|
14733
|
Nucleotide specificity of pyruvate kinase and phosphoenolpyruvate carboxykinase
|
|
14734
|
Nucleotide specificity of pyruvate kinase and phosphoenolpyruvate carboxykinase
|
|
14735
|
Nucleotide specificity of pyruvate kinase and phosphoenolpyruvate carboxykinase
|
|
14736
|
Nucleotide specificity of pyruvate kinase and phosphoenolpyruvate carboxykinase
|
|
14737
|
Nucleotide specificity of pyruvate kinase and phosphoenolpyruvate carboxykinase
|
|
14738
|
Nucleotide specificity of pyruvate kinase and phosphoenolpyruvate carboxykinase
|
|
14739
|
Nucleotide specificity of pyruvate kinase and phosphoenolpyruvate carboxykinase
|
|
14740
|
Nucleotide specificity of pyruvate kinase and phosphoenolpyruvate carboxykinase
|
|
14741
|
Nucleotide specificity of pyruvate kinase and phosphoenolpyruvate carboxykinase
|
|
14742
|
Nucleotide specificity of pyruvate kinase and phosphoenolpyruvate carboxykinase
|
|
14743
|
Nucleotide specificity of pyruvate kinase and phosphoenolpyruvate carboxykinase
|
|
14744
|
Nucleotide specificity of pyruvate kinase and phosphoenolpyruvate carboxykinase
|
|
14745
|
Nucleotide specificity of pyruvate kinase and phosphoenolpyruvate carboxykinase
|
|
14746
|
Nucleotide specificity of pyruvate kinase and phosphoenolpyruvate carboxykinase
|
|
14747
|
Nucleotide specificity of pyruvate kinase and phosphoenolpyruvate carboxykinase
|
|
14748
|
Nucleotide specificity of pyruvate kinase and phosphoenolpyruvate carboxykinase
|
|
14749
|
Nucleotide specificity of pyruvate kinase and phosphoenolpyruvate carboxykinase
|
|
14750
|
Nucleotide specificity of pyruvate kinase and phosphoenolpyruvate carboxykinase
|
|
14751
|
Nucleotide specificity of pyruvate kinase and phosphoenolpyruvate carboxykinase
|
|
14752
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14753
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14754
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14755
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14756
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14757
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14758
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14759
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14760
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14761
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14762
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14763
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14764
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14765
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14766
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14767
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14768
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14769
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14770
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14771
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14772
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14773
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14774
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14775
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14776
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14777
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14778
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14779
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14780
|
The effect of monovalent and divalent cations on the activity of Streptococcus lactis C10 pyruvate kinase
|
|
14781
|
Enzymatic characterization of a prokaryotic urea carboxylase
|
|
14782
|
Enzymatic characterization of a prokaryotic urea carboxylase
|
|
14783
|
Enzymatic characterization of a prokaryotic urea carboxylase
|
|
14784
|
Enzymatic characterization of a prokaryotic urea carboxylase
|
|
14785
|
Functional analysis of disease-causing mutations in human galactokinase
|
|
14786
|
Functional analysis of disease-causing mutations in human galactokinase
|
|
14787
|
Functional analysis of disease-causing mutations in human galactokinase
|
|
14788
|
Functional analysis of disease-causing mutations in human galactokinase
|
|
14789
|
Functional analysis of disease-causing mutations in human galactokinase
|
|
14790
|
Functional analysis of disease-causing mutations in human galactokinase
|
|
14791
|
Functional analysis of disease-causing mutations in human galactokinase
|
|
14792
|
Functional analysis of disease-causing mutations in human galactokinase
|
|
14793
|
Mutants of Escherichia coli defective in membrane phospholipid synthesis. Properties of wild type and Km ...
|
|
14794
|
Mutants of Escherichia coli defective in membrane phospholipid synthesis. Properties of wild type and Km ...
|
|
14795
|
Mutants of Escherichia coli defective in membrane phospholipid synthesis. Properties of wild type and Km ...
|
|
14796
|
Mutants of Escherichia coli defective in membrane phospholipid synthesis. Properties of wild type and Km ...
|
|
14797
|
Mutants of Escherichia coli defective in membrane phospholipid synthesis. Properties of wild type and Km ...
|
|
14798
|
Mutants of Escherichia coli defective in membrane phospholipid synthesis. Properties of wild type and Km ...
|
|
14799
|
Mutants of Escherichia coli defective in membrane phospholipid synthesis. Properties of wild type and Km ...
|
|
14800
|
Mutants of Escherichia coli defective in membrane phospholipid synthesis. Properties of wild type and Km ...
|
|
14801
|
Mutants of Escherichia coli defective in membrane phospholipid synthesis. Properties of wild type and Km ...
|
|
14802
|
Mutants of Escherichia coli defective in membrane phospholipid synthesis. Properties of wild type and Km ...
|
|
14803
|
Mutants of Escherichia coli defective in membrane phospholipid synthesis. Properties of wild type and Km ...
|
|
14804
|
Mutants of Escherichia coli defective in membrane phospholipid synthesis. Properties of wild type and Km ...
|
|
14805
|
Mutants of Escherichia coli defective in membrane phospholipid synthesis. Properties of wild type and Km ...
|
|
14806
|
Mutants of Escherichia coli defective in membrane phospholipid synthesis. Properties of wild type and Km ...
|
|
14807
|
Mutants of Escherichia coli defective in membrane phospholipid synthesis. Properties of wild type and Km ...
|
|
14808
|
Conserved cysteine residues of histidinol dehydrogenase are not involved in catalysis. Novel chemistry ...
|
|
14809
|
Conserved cysteine residues of histidinol dehydrogenase are not involved in catalysis. Novel chemistry ...
|
|
14810
|
Conserved cysteine residues of histidinol dehydrogenase are not involved in catalysis. Novel chemistry ...
|
|
14811
|
Conserved cysteine residues of histidinol dehydrogenase are not involved in catalysis. Novel chemistry ...
|
|
14812
|
Conserved cysteine residues of histidinol dehydrogenase are not involved in catalysis. Novel chemistry ...
|
|
14813
|
Conserved cysteine residues of histidinol dehydrogenase are not involved in catalysis. Novel chemistry ...
|
|
14814
|
Dominant Negative Role of the Glutamic Acid Residue Conserved in the Pyruvate Kinase M1 Isozyme in the ...
|
|
14815
|
Dominant Negative Role of the Glutamic Acid Residue Conserved in the Pyruvate Kinase M1 Isozyme in the ...
|
|
14816
|
Dominant Negative Role of the Glutamic Acid Residue Conserved in the Pyruvate Kinase M1 Isozyme in the ...
|
|
14817
|
Dominant Negative Role of the Glutamic Acid Residue Conserved in the Pyruvate Kinase M1 Isozyme in the ...
|
|
14818
|
Dominant Negative Role of the Glutamic Acid Residue Conserved in the Pyruvate Kinase M1 Isozyme in the ...
|
|
14819
|
Dominant Negative Role of the Glutamic Acid Residue Conserved in the Pyruvate Kinase M1 Isozyme in the ...
|
|
14820
|
Dominant Negative Role of the Glutamic Acid Residue Conserved in the Pyruvate Kinase M1 Isozyme in the ...
|
|
14821
|
Dominant Negative Role of the Glutamic Acid Residue Conserved in the Pyruvate Kinase M1 Isozyme in the ...
|
|
14822
|
Dominant Negative Role of the Glutamic Acid Residue Conserved in the Pyruvate Kinase M1 Isozyme in the ...
|
|
14823
|
Dominant Negative Role of the Glutamic Acid Residue Conserved in the Pyruvate Kinase M1 Isozyme in the ...
|
|
14824
|
Dominant Negative Role of the Glutamic Acid Residue Conserved in the Pyruvate Kinase M1 Isozyme in the ...
|
|
14825
|
Dominant Negative Role of the Glutamic Acid Residue Conserved in the Pyruvate Kinase M1 Isozyme in the ...
|
|
14826
|
Dominant Negative Role of the Glutamic Acid Residue Conserved in the Pyruvate Kinase M1 Isozyme in the ...
|
|
14827
|
Dominant Negative Role of the Glutamic Acid Residue Conserved in the Pyruvate Kinase M1 Isozyme in the ...
|
|
14828
|
Dominant Negative Role of the Glutamic Acid Residue Conserved in the Pyruvate Kinase M1 Isozyme in the ...
|
|
14829
|
Dominant Negative Role of the Glutamic Acid Residue Conserved in the Pyruvate Kinase M1 Isozyme in the ...
|
|
14830
|
Dominant Negative Role of the Glutamic Acid Residue Conserved in the Pyruvate Kinase M1 Isozyme in the ...
|
|
14831
|
Dominant Negative Role of the Glutamic Acid Residue Conserved in the Pyruvate Kinase M1 Isozyme in the ...
|
|
14832
|
Dominant Negative Role of the Glutamic Acid Residue Conserved in the Pyruvate Kinase M1 Isozyme in the ...
|
|
14833
|
Dominant Negative Role of the Glutamic Acid Residue Conserved in the Pyruvate Kinase M1 Isozyme in the ...
|
|
14834
|
Dominant Negative Role of the Glutamic Acid Residue Conserved in the Pyruvate Kinase M1 Isozyme in the ...
|
|
14835
|
Dominant Negative Role of the Glutamic Acid Residue Conserved in the Pyruvate Kinase M1 Isozyme in the ...
|
|
14836
|
Dominant Negative Role of the Glutamic Acid Residue Conserved in the Pyruvate Kinase M1 Isozyme in the ...
|
|
14837
|
Dominant Negative Role of the Glutamic Acid Residue Conserved in the Pyruvate Kinase M1 Isozyme in the ...
|
|
14838
|
IS981-mediated adaptive evolution recovers lactate production by ldhB transcription activation in a lactate ...
|
|
14839
|
IS981-mediated adaptive evolution recovers lactate production by ldhB transcription activation in a lactate ...
|
|
14840
|
IS981-mediated adaptive evolution recovers lactate production by ldhB transcription activation in a lactate ...
|
|
14841
|
IS981-mediated adaptive evolution recovers lactate production by ldhB transcription activation in a lactate ...
|
|
14842
|
IS981-mediated adaptive evolution recovers lactate production by ldhB transcription activation in a lactate ...
|
|
14843
|
IS981-mediated adaptive evolution recovers lactate production by ldhB transcription activation in a lactate ...
|
|
14844
|
Human cytosolic adenylate kinase allelozymes; purification and characterization
|
|
14845
|
Human cytosolic adenylate kinase allelozymes; purification and characterization
|
|
14846
|
Human cytosolic adenylate kinase allelozymes; purification and characterization
|
|
14847
|
Human cytosolic adenylate kinase allelozymes; purification and characterization
|
|
14848
|
Activation of yeast enolase by Cd(II)
|
|
14849
|
Activation of yeast enolase by Cd(II)
|
|
14850
|
Activation of yeast enolase by Cd(II)
|
|
14851
|
Activation of yeast enolase by Cd(II)
|
|
14852
|
Activation of yeast enolase by Cd(II)
|
|
14853
|
Activation of yeast enolase by Cd(II)
|
|
14854
|
Activation of yeast enolase by Cd(II)
|
|
14855
|
Activation of yeast enolase by Cd(II)
|
|
14862
|
Structural function of residue-209 in coumarin 7-hydroxylase (P450coh). Enzyme-kinetic studies and ...
|
|
14863
|
Structural function of residue-209 in coumarin 7-hydroxylase (P450coh). Enzyme-kinetic studies and ...
|
|
14864
|
Structural function of residue-209 in coumarin 7-hydroxylase (P450coh). Enzyme-kinetic studies and ...
|
|
14865
|
Structural function of residue-209 in coumarin 7-hydroxylase (P450coh). Enzyme-kinetic studies and ...
|
|
14866
|
Structural function of residue-209 in coumarin 7-hydroxylase (P450coh). Enzyme-kinetic studies and ...
|
|
14867
|
Structural function of residue-209 in coumarin 7-hydroxylase (P450coh). Enzyme-kinetic studies and ...
|
|
14868
|
Structural function of residue-209 in coumarin 7-hydroxylase (P450coh). Enzyme-kinetic studies and ...
|
|
14869
|
Structural function of residue-209 in coumarin 7-hydroxylase (P450coh). Enzyme-kinetic studies and ...
|
|
14870
|
Structural function of residue-209 in coumarin 7-hydroxylase (P450coh). Enzyme-kinetic studies and ...
|
|
14871
|
Structural function of residue-209 in coumarin 7-hydroxylase (P450coh). Enzyme-kinetic studies and ...
|
|
14872
|
Structural function of residue-209 in coumarin 7-hydroxylase (P450coh). Enzyme-kinetic studies and ...
|
|
14873
|
Structural function of residue-209 in coumarin 7-hydroxylase (P450coh). Enzyme-kinetic studies and ...
|
|
14874
|
Structural function of residue-209 in coumarin 7-hydroxylase (P450coh). Enzyme-kinetic studies and ...
|
|
14875
|
Structural function of residue-209 in coumarin 7-hydroxylase (P450coh). Enzyme-kinetic studies and ...
|
|
14876
|
Structural function of residue-209 in coumarin 7-hydroxylase (P450coh). Enzyme-kinetic studies and ...
|
|
14877
|
Structural function of residue-209 in coumarin 7-hydroxylase (P450coh). Enzyme-kinetic studies and ...
|
|
14878
|
Structural function of residue-209 in coumarin 7-hydroxylase (P450coh). Enzyme-kinetic studies and ...
|
|
14879
|
Structural function of residue-209 in coumarin 7-hydroxylase (P450coh). Enzyme-kinetic studies and ...
|
|
14880
|
Structural function of residue-209 in coumarin 7-hydroxylase (P450coh). Enzyme-kinetic studies and ...
|
|
14881
|
Functional characterization of an aminotransferase required for pyoverdine siderophore biosynthesis in ...
|
|
14882
|
Functional characterization of an aminotransferase required for pyoverdine siderophore biosynthesis in ...
|
|
14883
|
Structural and functional properties of aldose xylose reductase from the D-xylose-metabolizing yeast Candida ...
|
|
14884
|
Structural and functional properties of aldose xylose reductase from the D-xylose-metabolizing yeast Candida ...
|
|
14885
|
Structural and functional properties of aldose xylose reductase from the D-xylose-metabolizing yeast Candida ...
|
|
14886
|
Structural and functional properties of aldose xylose reductase from the D-xylose-metabolizing yeast Candida ...
|
|
14887
|
Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme ...
|
|
14888
|
Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme ...
|
|
14889
|
Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme ...
|
|
14890
|
Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme ...
|
|
14891
|
Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme ...
|
|
14892
|
Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme ...
|
|
14893
|
Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme ...
|
|
14894
|
Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme ...
|
|
14895
|
Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme ...
|
|
14896
|
Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme ...
|
|
14897
|
Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme ...
|
|
14898
|
Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme ...
|
|
14899
|
Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme ...
|
|
14900
|
Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme ...
|
|
14901
|
Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme ...
|
|
14902
|
Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme ...
|
|
14903
|
Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme ...
|
|
14904
|
Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme ...
|
|
14905
|
Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme ...
|
|
14906
|
Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme ...
|
|
14907
|
Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme ...
|
|
14908
|
Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme ...
|
|
14909
|
Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme ...
|
|
14910
|
Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme ...
|
|
14911
|
Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme ...
|
|
14912
|
Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme ...
|
|
14913
|
Identification of an Archaeal 2-Hydroxy Acid Dehydrogenase Catalyzing Reactions Involved in Coenzyme ...
|
|
14914
|
Competition of several enzymes for a common substrate: a possible model of cellular events
|
|
14915
|
Competition of several enzymes for a common substrate: a possible model of cellular events
|
|
14916
|
Competition of several enzymes for a common substrate: a possible model of cellular events
|
|
14917
|
Competition of several enzymes for a common substrate: a possible model of cellular events
|
|
14918
|
Competition of several enzymes for a common substrate: a possible model of cellular events
|
|
14919
|
Competition of several enzymes for a common substrate: a possible model of cellular events
|
|
14920
|
Competition of several enzymes for a common substrate: a possible model of cellular events
|
|
14921
|
Competition of several enzymes for a common substrate: a possible model of cellular events
|
|
14922
|
Competition of several enzymes for a common substrate: a possible model of cellular events
|
|
14923
|
Competition of several enzymes for a common substrate: a possible model of cellular events
|
|
14924
|
Competition of several enzymes for a common substrate: a possible model of cellular events
|
|
14925
|
Competition of several enzymes for a common substrate: a possible model of cellular events
|
|
14926
|
Competition of several enzymes for a common substrate: a possible model of cellular events
|
|
14927
|
Competition of several enzymes for a common substrate: a possible model of cellular events
|
|
14928
|
Competition of several enzymes for a common substrate: a possible model of cellular events
|
|
14929
|
Competition of several enzymes for a common substrate: a possible model of cellular events
|
|
14930
|
Competition of several enzymes for a common substrate: a possible model of cellular events
|
|
14931
|
Competition of several enzymes for a common substrate: a possible model of cellular events
|
|
14932
|
Competition of several enzymes for a common substrate: a possible model of cellular events
|
|
14933
|
Competition of several enzymes for a common substrate: a possible model of cellular events
|
|
14934
|
Competition of several enzymes for a common substrate: a possible model of cellular events
|
|
14935
|
Competition of several enzymes for a common substrate: a possible model of cellular events
|
|
14936
|
Competition of several enzymes for a common substrate: a possible model of cellular events
|
|
14937
|
Competition of several enzymes for a common substrate: a possible model of cellular events
|
|
14938
|
Competition of several enzymes for a common substrate: a possible model of cellular events
|
|
14939
|
Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and ...
|
|
14940
|
Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and ...
|
|
14941
|
Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and ...
|
|
14942
|
Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and ...
|
|
14943
|
Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and ...
|
|
14944
|
Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and ...
|
|
14945
|
Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and ...
|
|
14946
|
Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and ...
|
|
14947
|
Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and ...
|
|
14948
|
Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and ...
|
|
14949
|
Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and ...
|
|
14950
|
Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and ...
|
|
14951
|
Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and ...
|
|
14952
|
Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and ...
|
|
14953
|
Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and ...
|
|
14954
|
Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and ...
|
|
14955
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14956
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14957
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14958
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14959
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14960
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14961
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14962
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14963
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14964
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14965
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14966
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14967
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14968
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14969
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14970
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14971
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14972
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14973
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14974
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14975
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14976
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14977
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14978
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14979
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14980
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14981
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14982
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14983
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14984
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14985
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14986
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14987
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14988
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14989
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14990
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14991
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14992
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14993
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14994
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14995
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14996
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14997
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14998
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
14999
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|
|
15000
|
Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway ...
|





 If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.
If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.