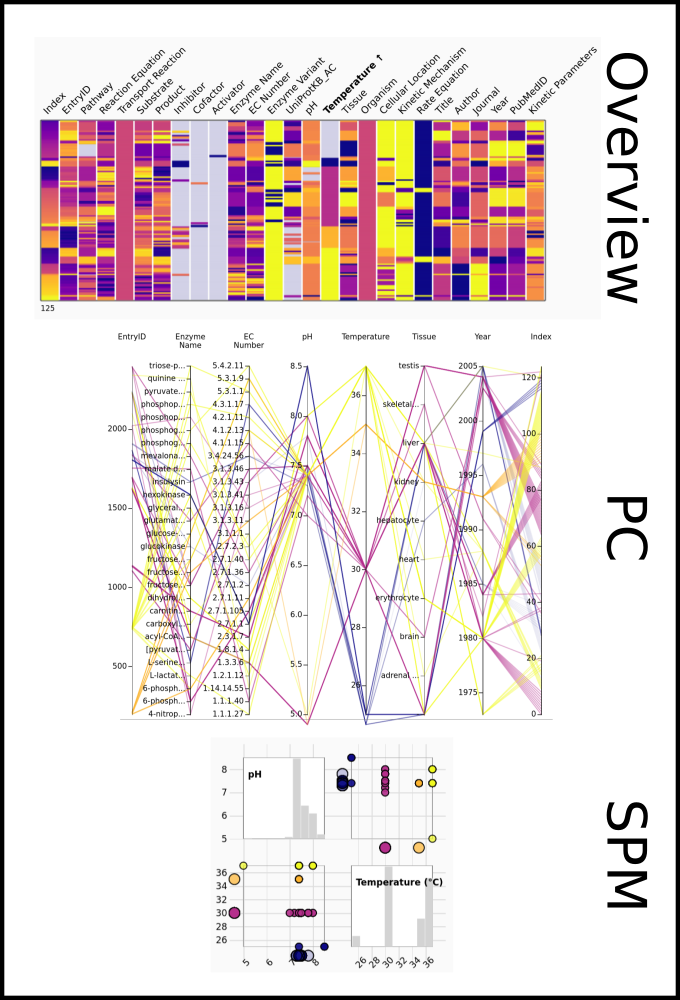
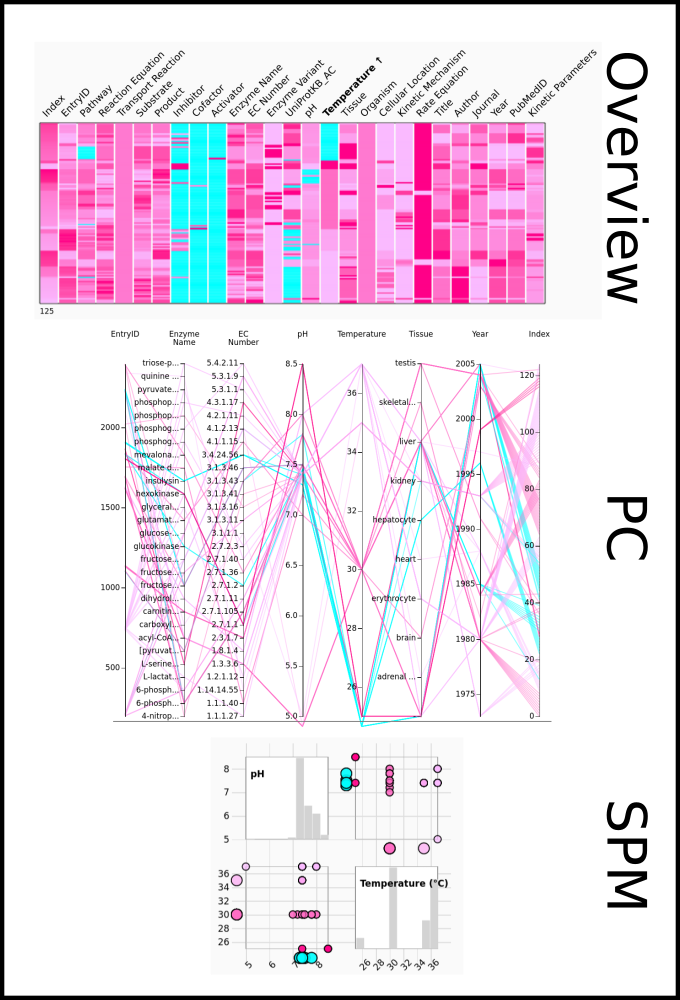
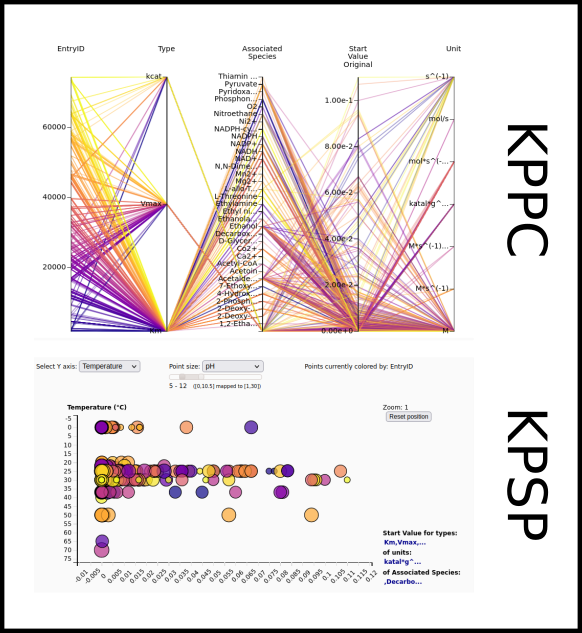
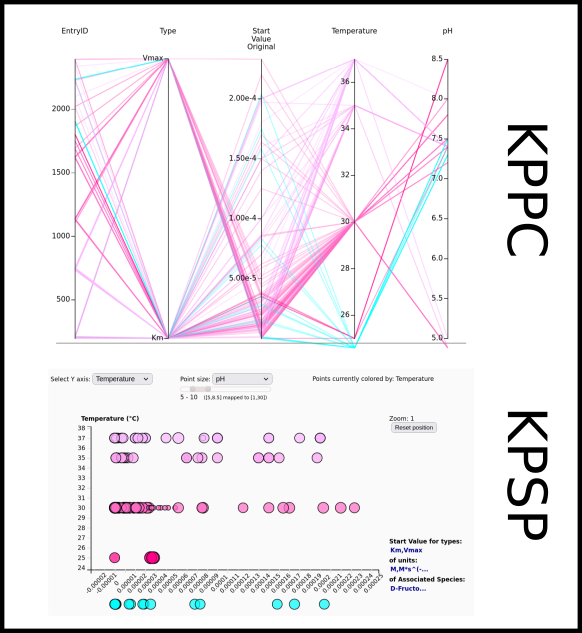
To improve SABIO-RK data interoperability, semantic markup was added to web pages as described and defined by
This structured information makes it easier to discover, collate and analyse our data.
|
11001
|
Kinetic characterization of mutations found in propionic acidemia and methylcrotonylglycinuria: evidence for ...
|
|
11002
|
Kinetic characterization of mutations found in propionic acidemia and methylcrotonylglycinuria: evidence for ...
|
|
11003
|
Kinetic characterization of mutations found in propionic acidemia and methylcrotonylglycinuria: evidence for ...
|
|
11004
|
Kinetic characterization of mutations found in propionic acidemia and methylcrotonylglycinuria: evidence for ...
|
|
11005
|
Kinetic characterization of mutations found in propionic acidemia and methylcrotonylglycinuria: evidence for ...
|
|
11006
|
Kinetic characterization of mutations found in propionic acidemia and methylcrotonylglycinuria: evidence for ...
|
|
11007
|
Kinetic characterization of mutations found in propionic acidemia and methylcrotonylglycinuria: evidence for ...
|
|
11008
|
Kinetic characterization of mutations found in propionic acidemia and methylcrotonylglycinuria: evidence for ...
|
|
11009
|
Kinetic characterization of mutations found in propionic acidemia and methylcrotonylglycinuria: evidence for ...
|
|
11010
|
Kinetic characterization of mutations found in propionic acidemia and methylcrotonylglycinuria: evidence for ...
|
|
11011
|
Kinetic characterization of mutations found in propionic acidemia and methylcrotonylglycinuria: evidence for ...
|
|
11012
|
Kinetic characterization of mutations found in propionic acidemia and methylcrotonylglycinuria: evidence for ...
|
|
11013
|
Kinetic characterization of mutations found in propionic acidemia and methylcrotonylglycinuria: evidence for ...
|
|
11014
|
Kinetic characterization of mutations found in propionic acidemia and methylcrotonylglycinuria: evidence for ...
|
|
11015
|
The prodrug activator EtaA from Mycobacterium tuberculosis is a Baeyer-Villiger monooxygenase
|
|
11016
|
The prodrug activator EtaA from Mycobacterium tuberculosis is a Baeyer-Villiger monooxygenase
|
|
11017
|
The prodrug activator EtaA from Mycobacterium tuberculosis is a Baeyer-Villiger monooxygenase
|
|
11018
|
The prodrug activator EtaA from Mycobacterium tuberculosis is a Baeyer-Villiger monooxygenase
|
|
11019
|
The prodrug activator EtaA from Mycobacterium tuberculosis is a Baeyer-Villiger monooxygenase
|
|
11020
|
Characterization of Coturnix quail liver alcohol dehydrogenase enzymes
|
|
11021
|
Characterization of Coturnix quail liver alcohol dehydrogenase enzymes
|
|
11022
|
Characterization of Coturnix quail liver alcohol dehydrogenase enzymes
|
|
11023
|
Characterization of Coturnix quail liver alcohol dehydrogenase enzymes
|
|
11024
|
Characterization of Coturnix quail liver alcohol dehydrogenase enzymes
|
|
11025
|
Characterization of Coturnix quail liver alcohol dehydrogenase enzymes
|
|
11026
|
Characterization of Coturnix quail liver alcohol dehydrogenase enzymes
|
|
11027
|
Characterization of Coturnix quail liver alcohol dehydrogenase enzymes
|
|
11028
|
Characterization of Coturnix quail liver alcohol dehydrogenase enzymes
|
|
11029
|
Characterization of Coturnix quail liver alcohol dehydrogenase enzymes
|
|
11030
|
Characterization of Coturnix quail liver alcohol dehydrogenase enzymes
|
|
11031
|
Characterization of Coturnix quail liver alcohol dehydrogenase enzymes
|
|
11032
|
Characterization of Coturnix quail liver alcohol dehydrogenase enzymes
|
|
11033
|
Do cysteine 230 and lysine 238 of biotin carboxylase play a role in the activation of biotin?
|
|
11034
|
Do cysteine 230 and lysine 238 of biotin carboxylase play a role in the activation of biotin?
|
|
11035
|
Do cysteine 230 and lysine 238 of biotin carboxylase play a role in the activation of biotin?
|
|
11036
|
Do cysteine 230 and lysine 238 of biotin carboxylase play a role in the activation of biotin?
|
|
11037
|
Do cysteine 230 and lysine 238 of biotin carboxylase play a role in the activation of biotin?
|
|
11038
|
Do cysteine 230 and lysine 238 of biotin carboxylase play a role in the activation of biotin?
|
|
11039
|
Do cysteine 230 and lysine 238 of biotin carboxylase play a role in the activation of biotin?
|
|
11040
|
Do cysteine 230 and lysine 238 of biotin carboxylase play a role in the activation of biotin?
|
|
11041
|
Do cysteine 230 and lysine 238 of biotin carboxylase play a role in the activation of biotin?
|
|
11042
|
Do cysteine 230 and lysine 238 of biotin carboxylase play a role in the activation of biotin?
|
|
11043
|
Do cysteine 230 and lysine 238 of biotin carboxylase play a role in the activation of biotin?
|
|
11044
|
Do cysteine 230 and lysine 238 of biotin carboxylase play a role in the activation of biotin?
|
|
11045
|
Do cysteine 230 and lysine 238 of biotin carboxylase play a role in the activation of biotin?
|
|
11046
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11047
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11048
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11049
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11050
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11051
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11052
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11053
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11054
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11055
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11056
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11057
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11058
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11059
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11060
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11061
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11062
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11063
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11064
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11065
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11066
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11067
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11068
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11069
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11070
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11071
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11072
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11073
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11074
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11075
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11076
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11077
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11078
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11079
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11080
|
Characterization of the Acyl Substrate Binding Pocket of Acetyl-CoA Synthetase
|
|
11081
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11082
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11083
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11084
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11085
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11086
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11087
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11088
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11089
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11090
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11091
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11092
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11093
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11094
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11095
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11096
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11097
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11098
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11099
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11100
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11101
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11102
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11103
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11104
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11105
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11106
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11107
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11108
|
Kinetic analysis of a new human ornithine carbamoyltransferase variant
|
|
11109
|
Purification and properties of NAD+-dependent glyceraldehyde-3-phosphate dehydrogenase from spinach leaves
|
|
11110
|
Purification and properties of NAD+-dependent glyceraldehyde-3-phosphate dehydrogenase from spinach leaves
|
|
11111
|
L-arginine binding to liver arginase requires proton transfer to gateway residue His141 and coordination of ...
|
|
11112
|
L-arginine binding to liver arginase requires proton transfer to gateway residue His141 and coordination of ...
|
|
11113
|
L-arginine binding to liver arginase requires proton transfer to gateway residue His141 and coordination of ...
|
|
11114
|
L-arginine binding to liver arginase requires proton transfer to gateway residue His141 and coordination of ...
|
|
11115
|
L-arginine binding to liver arginase requires proton transfer to gateway residue His141 and coordination of ...
|
|
11116
|
L-arginine binding to liver arginase requires proton transfer to gateway residue His141 and coordination of ...
|
|
11117
|
L-arginine binding to liver arginase requires proton transfer to gateway residue His141 and coordination of ...
|
|
11118
|
L-arginine binding to liver arginase requires proton transfer to gateway residue His141 and coordination of ...
|
|
11119
|
L-arginine binding to liver arginase requires proton transfer to gateway residue His141 and coordination of ...
|
|
11120
|
L-arginine binding to liver arginase requires proton transfer to gateway residue His141 and coordination of ...
|
|
11121
|
L-arginine binding to liver arginase requires proton transfer to gateway residue His141 and coordination of ...
|
|
11122
|
L-arginine binding to liver arginase requires proton transfer to gateway residue His141 and coordination of ...
|
|
11123
|
Wheatgerm hexokinase (LII): fluorimetric measurement of the binding of substrates and products
|
|
11124
|
Wheatgerm hexokinase (LII): fluorimetric measurement of the binding of substrates and products
|
|
11125
|
Wheatgerm hexokinase (LII): fluorimetric measurement of the binding of substrates and products
|
|
11126
|
Wheatgerm hexokinase (LII): fluorimetric measurement of the binding of substrates and products
|
|
11127
|
Wheatgerm hexokinase (LII): fluorimetric measurement of the binding of substrates and products
|
|
11128
|
Wheatgerm hexokinase (LII): fluorimetric measurement of the binding of substrates and products
|
|
11129
|
Cloning and expression of two secretory acetylcholinesterases from the bovine lungworm, Dictyocaulus viviparus
|
|
11130
|
Cloning and expression of two secretory acetylcholinesterases from the bovine lungworm, Dictyocaulus viviparus
|
|
11131
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11132
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11133
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11134
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11135
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11136
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11137
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11138
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11139
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11140
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11141
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11142
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11143
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11144
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11145
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11146
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11147
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11148
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11149
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11150
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11151
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11152
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11153
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11154
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11155
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11156
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11157
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11158
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11159
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11160
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11161
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11162
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11163
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11164
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11165
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11166
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11167
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11168
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11169
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11170
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11171
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11172
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11173
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11174
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11175
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11176
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11177
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11178
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11179
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11180
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11181
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11182
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11183
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11184
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11185
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11186
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11187
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11188
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11189
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11190
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11191
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11192
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11193
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11194
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11195
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11196
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11197
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11198
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11199
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11200
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11201
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11202
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11203
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11204
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11205
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11206
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11207
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11208
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11209
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11210
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11211
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11212
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11213
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11214
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11215
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11216
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11217
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11218
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11219
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11220
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11221
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11222
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11223
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11224
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11225
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11226
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11227
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11228
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11229
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11230
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11231
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11232
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11233
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11234
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11235
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11236
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11237
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11238
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11239
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11240
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11241
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11242
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11243
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11244
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11245
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11246
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11247
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11248
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11249
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11250
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11251
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11252
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11253
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11254
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11255
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11256
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11257
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11258
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11259
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11260
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11261
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11262
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11263
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11264
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11265
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11266
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11267
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11268
|
Molecular characterization of mammalian dicarbonyl/L-xylulose reductase and its localization in kidney
|
|
11269
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11270
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11271
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11272
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11273
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11274
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11275
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11276
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11277
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11278
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11279
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11280
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11281
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11282
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11283
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11284
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11285
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11286
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11287
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11288
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11289
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11290
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11291
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11292
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11293
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11294
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11295
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11296
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11297
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11298
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11299
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11300
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11301
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11302
|
Isolation and characterization of three alcohol dehydrogenase isozymes from Syrian golden hamsters
|
|
11303
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11304
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11305
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11306
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11307
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11308
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11309
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11310
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11311
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11312
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11313
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11314
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11315
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11316
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11317
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11318
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11319
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11320
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11321
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11322
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11323
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11324
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11325
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11326
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11327
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11328
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11329
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11330
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11331
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11332
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11333
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11334
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11335
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11336
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11337
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11338
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11339
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11340
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11341
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11342
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11343
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11344
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11345
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11346
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11347
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11348
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11349
|
The specificity of alcohol dehydrogenase with cis-retinoids. Activity with 11-cis-retinol and localization in ...
|
|
11350
|
Functional organization and evolution of mammalian hexokinases: mutations that caused the loss of catalytic ...
|
|
11351
|
Functional organization and evolution of mammalian hexokinases: mutations that caused the loss of catalytic ...
|
|
11352
|
Functional organization and evolution of mammalian hexokinases: mutations that caused the loss of catalytic ...
|
|
11353
|
Functional organization and evolution of mammalian hexokinases: mutations that caused the loss of catalytic ...
|
|
11354
|
Functional organization and evolution of mammalian hexokinases: mutations that caused the loss of catalytic ...
|
|
11355
|
Functional organization and evolution of mammalian hexokinases: mutations that caused the loss of catalytic ...
|
|
11356
|
Functional organization and evolution of mammalian hexokinases: mutations that caused the loss of catalytic ...
|
|
11357
|
Expression, crystallization and preliminary crystallographic analysis of human carbonyl reductase
|
|
11358
|
Expression, crystallization and preliminary crystallographic analysis of human carbonyl reductase
|
|
11359
|
Expression, crystallization and preliminary crystallographic analysis of human carbonyl reductase
|
|
11360
|
Expression, crystallization and preliminary crystallographic analysis of human carbonyl reductase
|
|
11361
|
Expression, crystallization and preliminary crystallographic analysis of human carbonyl reductase
|
|
11362
|
Expression, crystallization and preliminary crystallographic analysis of human carbonyl reductase
|
|
11363
|
Expression, crystallization and preliminary crystallographic analysis of human carbonyl reductase
|
|
11364
|
Expression, crystallization and preliminary crystallographic analysis of human carbonyl reductase
|
|
11365
|
Expression, crystallization and preliminary crystallographic analysis of human carbonyl reductase
|
|
11366
|
Expression, crystallization and preliminary crystallographic analysis of human carbonyl reductase
|
|
11367
|
Expression, crystallization and preliminary crystallographic analysis of human carbonyl reductase
|
|
11368
|
Substrate-dependent inhibition of yeast enolase by fluoride
|
|
11369
|
Purification and characterization of a new human prostatic acid phosphatase isoenzyme
|
|
11370
|
Purification and characterization of a new human prostatic acid phosphatase isoenzyme
|
|
11371
|
Purification and characterization of a new human prostatic acid phosphatase isoenzyme
|
|
11372
|
Purification and characterization of a new human prostatic acid phosphatase isoenzyme
|
|
11373
|
Purification and characterization of a new human prostatic acid phosphatase isoenzyme
|
|
11374
|
Purification and characterization of a new human prostatic acid phosphatase isoenzyme
|
|
11375
|
Purification and characterization of a new human prostatic acid phosphatase isoenzyme
|
|
11376
|
Purification and characterization of a new human prostatic acid phosphatase isoenzyme
|
|
11377
|
Purification and characterization of a new human prostatic acid phosphatase isoenzyme
|
|
11378
|
Purification and characterization of a new human prostatic acid phosphatase isoenzyme
|
|
11379
|
Purification and characterization of a new human prostatic acid phosphatase isoenzyme
|
|
11380
|
Purification and characterization of a new human prostatic acid phosphatase isoenzyme
|
|
11381
|
Allosteric and non-allosteric phosphofructokinases from Lactobacilli. Purification and properties of ...
|
|
11382
|
Allosteric and non-allosteric phosphofructokinases from Lactobacilli. Purification and properties of ...
|
|
11383
|
Allosteric and non-allosteric phosphofructokinases from Lactobacilli. Purification and properties of ...
|
|
11384
|
Allosteric and non-allosteric phosphofructokinases from Lactobacilli. Purification and properties of ...
|
|
11385
|
Allosteric and non-allosteric phosphofructokinases from Lactobacilli. Purification and properties of ...
|
|
11386
|
Allosteric and non-allosteric phosphofructokinases from Lactobacilli. Purification and properties of ...
|
|
11387
|
Allosteric and non-allosteric phosphofructokinases from Lactobacilli. Purification and properties of ...
|
|
11388
|
Allosteric and non-allosteric phosphofructokinases from Lactobacilli. Purification and properties of ...
|
|
11389
|
Allosteric and non-allosteric phosphofructokinases from Lactobacilli. Purification and properties of ...
|
|
11390
|
Allosteric and non-allosteric phosphofructokinases from Lactobacilli. Purification and properties of ...
|
|
11391
|
Allosteric and non-allosteric phosphofructokinases from Lactobacilli. Purification and properties of ...
|
|
11392
|
Allosteric and non-allosteric phosphofructokinases from Lactobacilli. Purification and properties of ...
|
|
11393
|
Allosteric and non-allosteric phosphofructokinases from Lactobacilli. Purification and properties of ...
|
|
11394
|
Allosteric and non-allosteric phosphofructokinases from Lactobacilli. Purification and properties of ...
|
|
11395
|
Allosteric and non-allosteric phosphofructokinases from Lactobacilli. Purification and properties of ...
|
|
11396
|
Allosteric and non-allosteric phosphofructokinases from Lactobacilli. Purification and properties of ...
|
|
11397
|
Purification and characterization of NADPH-dependent carbonyl reductase, involved in stereoselective reduction ...
|
|
11398
|
Purification and characterization of NADPH-dependent carbonyl reductase, involved in stereoselective reduction ...
|
|
11399
|
Purification and characterization of NADPH-dependent carbonyl reductase, involved in stereoselective reduction ...
|
|
11400
|
Purification and characterization of NADPH-dependent carbonyl reductase, involved in stereoselective reduction ...
|
|
11401
|
Purification and characterization of NADPH-dependent carbonyl reductase, involved in stereoselective reduction ...
|
|
11402
|
Purification and characterization of NADPH-dependent carbonyl reductase, involved in stereoselective reduction ...
|
|
11403
|
Purification and characterization of NADPH-dependent carbonyl reductase, involved in stereoselective reduction ...
|
|
11404
|
Purification and characterization of NADPH-dependent carbonyl reductase, involved in stereoselective reduction ...
|
|
11405
|
Purification and characterization of NADPH-dependent carbonyl reductase, involved in stereoselective reduction ...
|
|
11406
|
Purification and characterization of NADPH-dependent carbonyl reductase, involved in stereoselective reduction ...
|
|
11407
|
Purification and characterization of NADPH-dependent carbonyl reductase, involved in stereoselective reduction ...
|
|
11408
|
Purification of human hepatic arginase and its manganese (II)-dependent and pH-dependent interconversion ...
|
|
11409
|
Purification of human hepatic arginase and its manganese (II)-dependent and pH-dependent interconversion ...
|
|
11410
|
Purification of human hepatic arginase and its manganese (II)-dependent and pH-dependent interconversion ...
|
|
11411
|
Purification of human hepatic arginase and its manganese (II)-dependent and pH-dependent interconversion ...
|
|
11412
|
Probing domain mobility in a flavocytochrome
|
|
11413
|
Probing domain mobility in a flavocytochrome
|
|
11414
|
Probing domain mobility in a flavocytochrome
|
|
11415
|
Probing domain mobility in a flavocytochrome
|
|
11416
|
Probing domain mobility in a flavocytochrome
|
|
11417
|
Probing domain mobility in a flavocytochrome
|
|
11418
|
Probing domain mobility in a flavocytochrome
|
|
11419
|
Probing domain mobility in a flavocytochrome
|
|
11420
|
Probing domain mobility in a flavocytochrome
|
|
11421
|
Probing domain mobility in a flavocytochrome
|
|
11422
|
Probing domain mobility in a flavocytochrome
|
|
11423
|
Probing domain mobility in a flavocytochrome
|
|
11424
|
Probing domain mobility in a flavocytochrome
|
|
11425
|
Probing domain mobility in a flavocytochrome
|
|
11426
|
Probing domain mobility in a flavocytochrome
|
|
11427
|
Kinetic studies dealing with immobilized reversible enzyme system. Experimental evidence for shift of apparent ...
|
|
11428
|
Kinetic studies dealing with immobilized reversible enzyme system. Experimental evidence for shift of apparent ...
|
|
11429
|
Evolution of Enzymatic Activities in the Enolase Superfamily: d-Tartrate Dehydratase from Bradyrhizobium ...
|
|
11430
|
Evolution of Enzymatic Activities in the Enolase Superfamily: d-Tartrate Dehydratase from Bradyrhizobium ...
|
|
11431
|
Evolution of Enzymatic Activities in the Enolase Superfamily: d-Tartrate Dehydratase from Bradyrhizobium ...
|
|
11432
|
Evolution of Enzymatic Activities in the Enolase Superfamily: d-Tartrate Dehydratase from Bradyrhizobium ...
|
|
11433
|
Evolution of Enzymatic Activities in the Enolase Superfamily: d-Tartrate Dehydratase from Bradyrhizobium ...
|
|
11434
|
Evolution of Enzymatic Activities in the Enolase Superfamily: d-Tartrate Dehydratase from Bradyrhizobium ...
|
|
11455
|
Isozymes of human phosphofructokinase: identification and subunit structural characterization of a new system
|
|
11456
|
Isozymes of human phosphofructokinase: identification and subunit structural characterization of a new system
|
|
11457
|
Isozymes of human phosphofructokinase: identification and subunit structural characterization of a new system
|
|
11458
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11459
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11460
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11461
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11462
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11463
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11464
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11465
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11466
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11467
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11468
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11469
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11470
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11471
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11472
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11473
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11474
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11475
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11476
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11477
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11478
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11479
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11480
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11481
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11482
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11483
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11484
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11485
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11486
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11487
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11488
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11489
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11490
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11491
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11492
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11493
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11494
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11495
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11496
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11497
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11498
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11499
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11500
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11501
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11502
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11503
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11504
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11505
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11506
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11507
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11508
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11509
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11510
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11511
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11512
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11513
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11514
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11515
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11516
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11517
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11518
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11519
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11520
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11521
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11522
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11523
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11524
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11525
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11526
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11527
|
Dissection of the human acetylcholinesterase active center determinants of substrate specificity. ...
|
|
11528
|
Substrate and inhibitor specificities of the thermostable alcohol dehydrogenase allozymes ADH-71k and ...
|
|
11529
|
Substrate and inhibitor specificities of the thermostable alcohol dehydrogenase allozymes ADH-71k and ...
|
|
11530
|
Substrate and inhibitor specificities of the thermostable alcohol dehydrogenase allozymes ADH-71k and ...
|
|
11531
|
Substrate and inhibitor specificities of the thermostable alcohol dehydrogenase allozymes ADH-71k and ...
|
|
11532
|
Substrate and inhibitor specificities of the thermostable alcohol dehydrogenase allozymes ADH-71k and ...
|
|
11533
|
Substrate and inhibitor specificities of the thermostable alcohol dehydrogenase allozymes ADH-71k and ...
|
|
11534
|
Multiple forms of Drosophila hexokinase. Purification, biochemical and immunological characterization
|
|
11535
|
Multiple forms of Drosophila hexokinase. Purification, biochemical and immunological characterization
|
|
11536
|
Multiple forms of Drosophila hexokinase. Purification, biochemical and immunological characterization
|
|
11537
|
Multiple forms of Drosophila hexokinase. Purification, biochemical and immunological characterization
|
|
11538
|
Multiple forms of Drosophila hexokinase. Purification, biochemical and immunological characterization
|
|
11539
|
Multiple forms of Drosophila hexokinase. Purification, biochemical and immunological characterization
|
|
11540
|
Multiple forms of Drosophila hexokinase. Purification, biochemical and immunological characterization
|
|
11541
|
Multiple forms of Drosophila hexokinase. Purification, biochemical and immunological characterization
|
|
11542
|
Multiple forms of Drosophila hexokinase. Purification, biochemical and immunological characterization
|
|
11543
|
Multiple forms of Drosophila hexokinase. Purification, biochemical and immunological characterization
|
|
11544
|
Multiple forms of Drosophila hexokinase. Purification, biochemical and immunological characterization
|
|
11545
|
Multiple forms of Drosophila hexokinase. Purification, biochemical and immunological characterization
|
|
11546
|
Regulation of energy metabolism in Trypanosoma (Schizotrypanum) cruzi epimastigotes. I. Hexokinase and ...
|
|
11547
|
Regulation of energy metabolism in Trypanosoma (Schizotrypanum) cruzi epimastigotes. I. Hexokinase and ...
|
|
11548
|
Regulation of energy metabolism in Trypanosoma (Schizotrypanum) cruzi epimastigotes. I. Hexokinase and ...
|
|
11549
|
Regulation of energy metabolism in Trypanosoma (Schizotrypanum) cruzi epimastigotes. I. Hexokinase and ...
|
|
11550
|
Regulation of energy metabolism in Trypanosoma (Schizotrypanum) cruzi epimastigotes. I. Hexokinase and ...
|
|
11551
|
Regulation of energy metabolism in Trypanosoma (Schizotrypanum) cruzi epimastigotes. I. Hexokinase and ...
|
|
11552
|
Regulation of energy metabolism in Trypanosoma (Schizotrypanum) cruzi epimastigotes. I. Hexokinase and ...
|
|
11553
|
Regulation of energy metabolism in Trypanosoma (Schizotrypanum) cruzi epimastigotes. I. Hexokinase and ...
|
|
11554
|
Regulation of energy metabolism in Trypanosoma (Schizotrypanum) cruzi epimastigotes. I. Hexokinase and ...
|
|
11555
|
Regulation of energy metabolism in Trypanosoma (Schizotrypanum) cruzi epimastigotes. I. Hexokinase and ...
|
|
11556
|
Regulation of energy metabolism in Trypanosoma (Schizotrypanum) cruzi epimastigotes. I. Hexokinase and ...
|
|
11557
|
Regulation of energy metabolism in Trypanosoma (Schizotrypanum) cruzi epimastigotes. I. Hexokinase and ...
|
|
11558
|
Regulation of energy metabolism in Trypanosoma (Schizotrypanum) cruzi epimastigotes. I. Hexokinase and ...
|
|
11559
|
A cadmium-binding protein in rat liver identified as ornithine carbamoyltransferase
|
|
11560
|
A cadmium-binding protein in rat liver identified as ornithine carbamoyltransferase
|
|
11561
|
A cadmium-binding protein in rat liver identified as ornithine carbamoyltransferase
|
|
11562
|
Isolation and characterization of ornithine transcarbamylase from normal human liver
|
|
11563
|
Isolation and characterization of ornithine transcarbamylase from normal human liver
|
|
11564
|
S-Adenosylhomocysteine hydrolase from hamster liver: purification and kinetic properties
|
|
11565
|
S-Adenosylhomocysteine hydrolase from hamster liver: purification and kinetic properties
|
|
11566
|
S-Adenosylhomocysteine hydrolase from hamster liver: purification and kinetic properties
|
|
11567
|
S-Adenosylhomocysteine hydrolase from hamster liver: purification and kinetic properties
|
|
11568
|
S-Adenosylhomocysteine hydrolase from hamster liver: purification and kinetic properties
|
|
11569
|
S-Adenosylhomocysteine hydrolase from hamster liver: purification and kinetic properties
|
|
11570
|
S-Adenosylhomocysteine hydrolase from hamster liver: purification and kinetic properties
|
|
11571
|
S-Adenosylhomocysteine hydrolase from hamster liver: purification and kinetic properties
|
|
11572
|
S-Adenosylhomocysteine hydrolase from hamster liver: purification and kinetic properties
|
|
11573
|
S-Adenosylhomocysteine hydrolase from hamster liver: purification and kinetic properties
|
|
11574
|
S-Adenosylhomocysteine hydrolase from hamster liver: purification and kinetic properties
|
|
11575
|
S-Adenosylhomocysteine hydrolase from hamster liver: purification and kinetic properties
|
|
11576
|
The chemical mechanism of action of glucose oxidase from Aspergillus niger
|
|
11577
|
The chemical mechanism of action of glucose oxidase from Aspergillus niger
|
|
11578
|
The chemical mechanism of action of glucose oxidase from Aspergillus niger
|
|
11579
|
The chemical mechanism of action of glucose oxidase from Aspergillus niger
|
|
11580
|
The chemical mechanism of action of glucose oxidase from Aspergillus niger
|
|
11581
|
The chemical mechanism of action of glucose oxidase from Aspergillus niger
|
|
11582
|
The chemical mechanism of action of glucose oxidase from Aspergillus niger
|
|
11583
|
The chemical mechanism of action of glucose oxidase from Aspergillus niger
|
|
11584
|
The chemical mechanism of action of glucose oxidase from Aspergillus niger
|
|
11585
|
The chemical mechanism of action of glucose oxidase from Aspergillus niger
|
|
11586
|
The chemical mechanism of action of glucose oxidase from Aspergillus niger
|
|
11587
|
The chemical mechanism of action of glucose oxidase from Aspergillus niger
|
|
11588
|
The chemical mechanism of action of glucose oxidase from Aspergillus niger
|
|
11589
|
The chemical mechanism of action of glucose oxidase from Aspergillus niger
|
|
11590
|
The chemical mechanism of action of glucose oxidase from Aspergillus niger
|
|
11591
|
The chemical mechanism of action of glucose oxidase from Aspergillus niger
|
|
11592
|
The chemical mechanism of action of glucose oxidase from Aspergillus niger
|
|
11593
|
The chemical mechanism of action of glucose oxidase from Aspergillus niger
|
|
11594
|
Zinc, a novel structural element found in the family of bacterial adenylate kinases
|
|
11595
|
Zinc, a novel structural element found in the family of bacterial adenylate kinases
|
|
11596
|
Zinc, a novel structural element found in the family of bacterial adenylate kinases
|
|
11597
|
Zinc, a novel structural element found in the family of bacterial adenylate kinases
|
|
11598
|
Zinc, a novel structural element found in the family of bacterial adenylate kinases
|
|
11599
|
Zinc, a novel structural element found in the family of bacterial adenylate kinases
|
|
11600
|
Zinc, a novel structural element found in the family of bacterial adenylate kinases
|
|
11601
|
Zinc, a novel structural element found in the family of bacterial adenylate kinases
|
|
11602
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11603
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11604
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11605
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11606
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11607
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11608
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11609
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11610
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11611
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11612
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11613
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11614
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11615
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11616
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11617
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11618
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11619
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11620
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11621
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11622
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11623
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11624
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11625
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11626
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11627
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11628
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11629
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11630
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11631
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11632
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11633
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11634
|
The role of hydrophobic active-site residues in substrate specificity and acyl transfer activity of penicillin ...
|
|
11635
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11636
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11637
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11638
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11639
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11640
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11641
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11642
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11643
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11644
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11645
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11646
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11647
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11648
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11649
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11650
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11651
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11652
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11653
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11654
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11655
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11656
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11657
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11658
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11659
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11660
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11661
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11662
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11663
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11664
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11665
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11666
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11667
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11668
|
Analysis of the substrate specificity of human Sulfotransferases SULT1A1 and SULT1A3: Site-directed ...
|
|
11669
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11670
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11671
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11672
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11673
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11674
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11675
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11676
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11677
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11678
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11679
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11680
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11681
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11682
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11683
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11684
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11685
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11686
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11687
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11688
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11689
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11690
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11691
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11692
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11693
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11694
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11695
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11696
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11697
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11698
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11699
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11700
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11701
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11702
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11703
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11704
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11705
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11706
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11707
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11708
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11709
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11710
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11711
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11712
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11713
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11714
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11715
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11716
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11717
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11718
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11719
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11720
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11721
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11722
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11723
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11724
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11725
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11726
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11727
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11728
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11729
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11730
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11731
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11732
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11733
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11734
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11735
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11736
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11737
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11738
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11739
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11740
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11741
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11742
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11743
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11744
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11745
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11746
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11747
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11748
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11749
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11750
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11751
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11752
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11753
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11754
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11755
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11756
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11757
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11758
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11759
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11760
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11761
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11762
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11763
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11764
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11765
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11766
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11767
|
Probing substrate binding to metallo-beta-lactamase L1 from Stenotrophomonas maltophilia by using ...
|
|
11768
|
Identification of Amino Acid Residues Involved in the Activity of Phosphomannose Isomerase-Guanosine ...
|
|
11769
|
Identification of Amino Acid Residues Involved in the Activity of Phosphomannose Isomerase-Guanosine ...
|
|
11770
|
Identification of Amino Acid Residues Involved in the Activity of Phosphomannose Isomerase-Guanosine ...
|
|
11771
|
Identification of Amino Acid Residues Involved in the Activity of Phosphomannose Isomerase-Guanosine ...
|
|
11772
|
Identification of Amino Acid Residues Involved in the Activity of Phosphomannose Isomerase-Guanosine ...
|
|
11773
|
Identification of Amino Acid Residues Involved in the Activity of Phosphomannose Isomerase-Guanosine ...
|
|
11774
|
Identification of Amino Acid Residues Involved in the Activity of Phosphomannose Isomerase-Guanosine ...
|
|
11775
|
Identification of Amino Acid Residues Involved in the Activity of Phosphomannose Isomerase-Guanosine ...
|
|
11776
|
Identification of Amino Acid Residues Involved in the Activity of Phosphomannose Isomerase-Guanosine ...
|
|
11777
|
Identification of Amino Acid Residues Involved in the Activity of Phosphomannose Isomerase-Guanosine ...
|
|
11778
|
Identification of Amino Acid Residues Involved in the Activity of Phosphomannose Isomerase-Guanosine ...
|
|
11779
|
Identification of Amino Acid Residues Involved in the Activity of Phosphomannose Isomerase-Guanosine ...
|
|
11780
|
Identification of Amino Acid Residues Involved in the Activity of Phosphomannose Isomerase-Guanosine ...
|
|
11781
|
Identification of Amino Acid Residues Involved in the Activity of Phosphomannose Isomerase-Guanosine ...
|
|
11782
|
Identification of Amino Acid Residues Involved in the Activity of Phosphomannose Isomerase-Guanosine ...
|
|
11783
|
Identification of Amino Acid Residues Involved in the Activity of Phosphomannose Isomerase-Guanosine ...
|
|
11784
|
Identification of Amino Acid Residues Involved in the Activity of Phosphomannose Isomerase-Guanosine ...
|
|
11785
|
Identification of Amino Acid Residues Involved in the Activity of Phosphomannose Isomerase-Guanosine ...
|
|
11786
|
Simian liver alcohol dehydrogenase: isolation and characterization of isoenzymes from Saimiri sciureus
|
|
11787
|
Simian liver alcohol dehydrogenase: isolation and characterization of isoenzymes from Saimiri sciureus
|
|
11788
|
Simian liver alcohol dehydrogenase: isolation and characterization of isoenzymes from Saimiri sciureus
|
|
11789
|
Simian liver alcohol dehydrogenase: isolation and characterization of isoenzymes from Saimiri sciureus
|
|
11790
|
Simian liver alcohol dehydrogenase: isolation and characterization of isoenzymes from Saimiri sciureus
|
|
11791
|
Simian liver alcohol dehydrogenase: isolation and characterization of isoenzymes from Saimiri sciureus
|
|
11792
|
Simian liver alcohol dehydrogenase: isolation and characterization of isoenzymes from Saimiri sciureus
|
|
11793
|
Simian liver alcohol dehydrogenase: isolation and characterization of isoenzymes from Saimiri sciureus
|
|
11794
|
Simian liver alcohol dehydrogenase: isolation and characterization of isoenzymes from Saimiri sciureus
|
|
11795
|
Simian liver alcohol dehydrogenase: isolation and characterization of isoenzymes from Saimiri sciureus
|
|
11796
|
Simian liver alcohol dehydrogenase: isolation and characterization of isoenzymes from Saimiri sciureus
|
|
11797
|
Simian liver alcohol dehydrogenase: isolation and characterization of isoenzymes from Saimiri sciureus
|
|
11798
|
Simian liver alcohol dehydrogenase: isolation and characterization of isoenzymes from Saimiri sciureus
|
|
11799
|
Simian liver alcohol dehydrogenase: isolation and characterization of isoenzymes from Saimiri sciureus
|
|
11800
|
Simian liver alcohol dehydrogenase: isolation and characterization of isoenzymes from Saimiri sciureus
|
|
11801
|
Simian liver alcohol dehydrogenase: isolation and characterization of isoenzymes from Saimiri sciureus
|
|
11802
|
Simian liver alcohol dehydrogenase: isolation and characterization of isoenzymes from Saimiri sciureus
|
|
11803
|
Simian liver alcohol dehydrogenase: isolation and characterization of isoenzymes from Saimiri sciureus
|
|
11804
|
Dual functional roles of ATP in the human mitochondrial malic enzyme
|
|
11805
|
Dual functional roles of ATP in the human mitochondrial malic enzyme
|
|
11806
|
Dual functional roles of ATP in the human mitochondrial malic enzyme
|
|
11807
|
Dual functional roles of ATP in the human mitochondrial malic enzyme
|
|
11808
|
Dual functional roles of ATP in the human mitochondrial malic enzyme
|
|
11809
|
Dual functional roles of ATP in the human mitochondrial malic enzyme
|
|
11810
|
Dual functional roles of ATP in the human mitochondrial malic enzyme
|
|
11811
|
Dual functional roles of ATP in the human mitochondrial malic enzyme
|
|
11812
|
Dual functional roles of ATP in the human mitochondrial malic enzyme
|
|
11813
|
Dual functional roles of ATP in the human mitochondrial malic enzyme
|
|
11814
|
Dual functional roles of ATP in the human mitochondrial malic enzyme
|
|
11815
|
Dual functional roles of ATP in the human mitochondrial malic enzyme
|
|
11816
|
Dual functional roles of ATP in the human mitochondrial malic enzyme
|
|
11817
|
Dual functional roles of ATP in the human mitochondrial malic enzyme
|
|
11818
|
Dual functional roles of ATP in the human mitochondrial malic enzyme
|
|
11819
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11820
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11821
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11822
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11823
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11824
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11825
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11826
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11827
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11828
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11829
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11830
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11831
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11832
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11833
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11834
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11835
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11836
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11837
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11838
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11839
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11840
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11841
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11842
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11843
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11844
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11845
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11846
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11847
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11848
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11849
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11850
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11851
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11852
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11853
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11854
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11855
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11856
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11857
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11858
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11859
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11860
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11861
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11862
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11863
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11864
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11865
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11866
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11867
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11868
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11869
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11870
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11871
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11872
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11873
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11874
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11875
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11876
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11877
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11878
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11879
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11880
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11881
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11882
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11883
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11884
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11885
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11886
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11887
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11888
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11889
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11890
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11891
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11892
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11893
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11894
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11895
|
Characterization of heterodimeric alkaline phosphatases from Escherichia coli: an investigation of intragenic ...
|
|
11896
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11897
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11898
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11899
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11900
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11901
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11902
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11903
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11904
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11905
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11906
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11907
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11908
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11909
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11910
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11911
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11912
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11913
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11914
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11915
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11916
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11917
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11918
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11919
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11920
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11921
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11922
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11923
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11924
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11925
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11926
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11927
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11928
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11929
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11930
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11931
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11932
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11933
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11934
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11935
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11936
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11937
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11938
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11939
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11940
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11941
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11942
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11943
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11944
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11945
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11946
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11947
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11948
|
Kinetic properties of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. Evidence for the ...
|
|
11949
|
Substrate diversity of herpes simplex virus thymidine kinase. Impact Of the kinematics of the enzyme
|
|
11950
|
Substrate diversity of herpes simplex virus thymidine kinase. Impact Of the kinematics of the enzyme
|
|
11951
|
Substrate diversity of herpes simplex virus thymidine kinase. Impact Of the kinematics of the enzyme
|
|
11952
|
Substrate diversity of herpes simplex virus thymidine kinase. Impact Of the kinematics of the enzyme
|
|
11953
|
Substrate diversity of herpes simplex virus thymidine kinase. Impact Of the kinematics of the enzyme
|
|
11954
|
Substrate diversity of herpes simplex virus thymidine kinase. Impact Of the kinematics of the enzyme
|
|
11955
|
Substrate diversity of herpes simplex virus thymidine kinase. Impact Of the kinematics of the enzyme
|
|
11956
|
Substrate diversity of herpes simplex virus thymidine kinase. Impact Of the kinematics of the enzyme
|
|
11957
|
Substrate diversity of herpes simplex virus thymidine kinase. Impact Of the kinematics of the enzyme
|
|
11958
|
Substrate diversity of herpes simplex virus thymidine kinase. Impact Of the kinematics of the enzyme
|
|
11959
|
Purification and Characterization of Acetate Kinase from Acetate-grown Methanosarcina thermophila- EVIDENCE ...
|
|
11960
|
Purification and Characterization of Acetate Kinase from Acetate-grown Methanosarcina thermophila- EVIDENCE ...
|
|
11961
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11962
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11963
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11964
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11965
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11966
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11967
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11968
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11969
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11970
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11971
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11972
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11973
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11974
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11975
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11976
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11977
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11978
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11979
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11980
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11981
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11982
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11983
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11984
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11985
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11986
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11987
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11988
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11989
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11990
|
Purification and characterisation of a lactococcal aminoacylase
|
|
11991
|
A new case of phosphoglycerate kinase deficiency: PGK Creteil associated with rhabdomyolysis and lacking ...
|
|
11992
|
A new case of phosphoglycerate kinase deficiency: PGK Creteil associated with rhabdomyolysis and lacking ...
|
|
11993
|
A new case of phosphoglycerate kinase deficiency: PGK Creteil associated with rhabdomyolysis and lacking ...
|
|
11994
|
A new case of phosphoglycerate kinase deficiency: PGK Creteil associated with rhabdomyolysis and lacking ...
|
|
11995
|
A new case of phosphoglycerate kinase deficiency: PGK Creteil associated with rhabdomyolysis and lacking ...
|
|
11996
|
A new case of phosphoglycerate kinase deficiency: PGK Creteil associated with rhabdomyolysis and lacking ...
|
|
11997
|
A new case of phosphoglycerate kinase deficiency: PGK Creteil associated with rhabdomyolysis and lacking ...
|
|
11998
|
A new case of phosphoglycerate kinase deficiency: PGK Creteil associated with rhabdomyolysis and lacking ...
|
|
11999
|
N5-(L-1-carboxyethyl)-L-ornithine:NADP+ oxidoreductase from Streptococcus lactis. Purification and partial ...
|
|
12000
|
N5-(L-1-carboxyethyl)-L-ornithine:NADP+ oxidoreductase from Streptococcus lactis. Purification and partial ...
|





 If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.
If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.