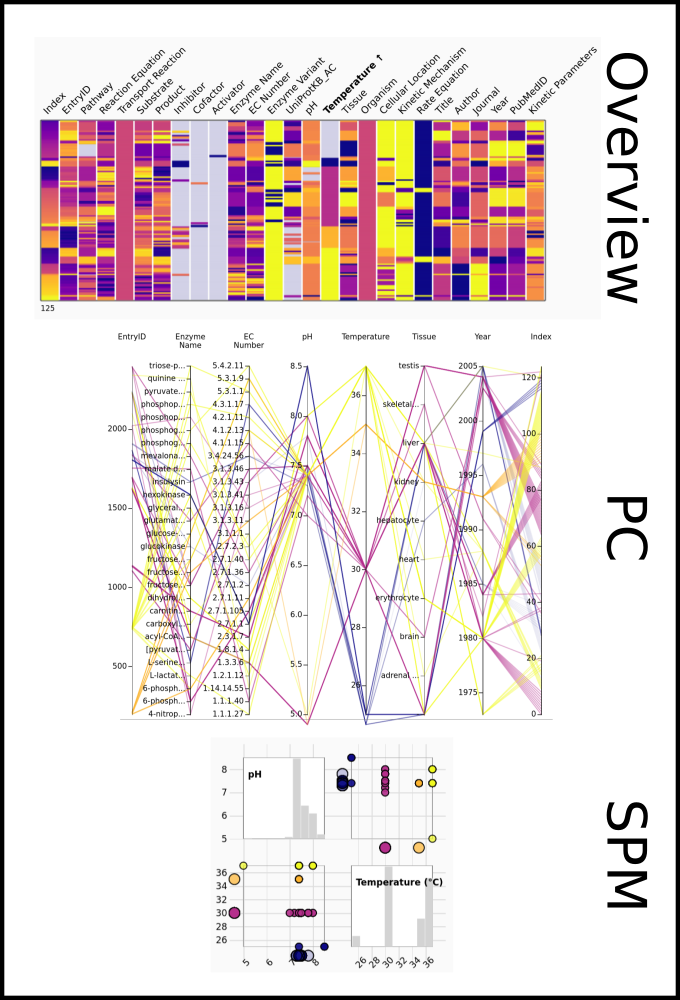
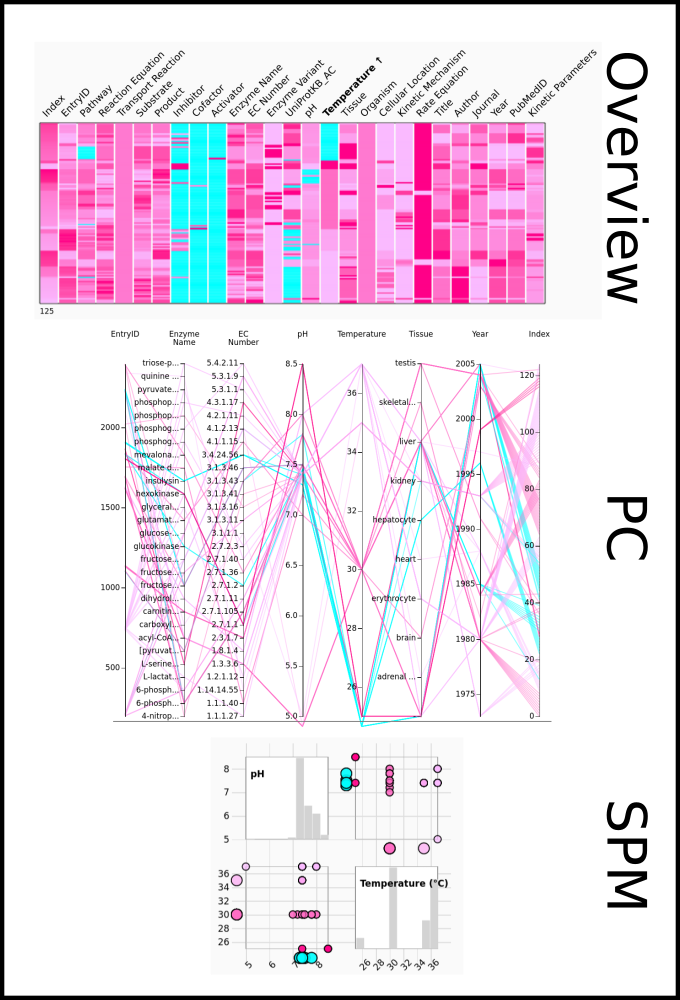
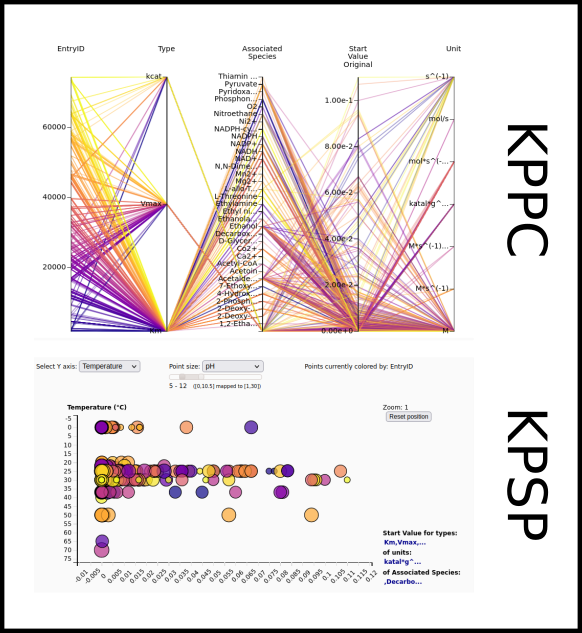
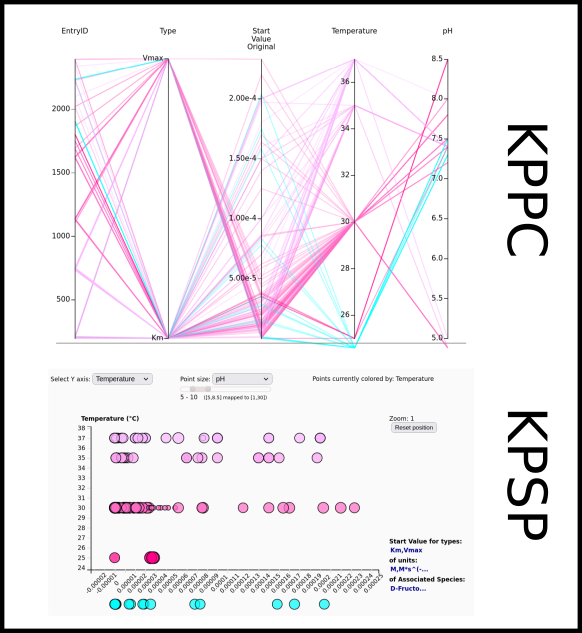
To improve SABIO-RK data interoperability, semantic markup was added to web pages as described and defined by
This structured information makes it easier to discover, collate and analyse our data.
|
1
|
Do enzymes change the nature of transition states? Mapping the transition state for general acid-base ...
|
|
2
|
Do enzymes change the nature of transition states? Mapping the transition state for general acid-base ...
|
|
3
|
Do enzymes change the nature of transition states? Mapping the transition state for general acid-base ...
|
|
4
|
Do enzymes change the nature of transition states? Mapping the transition state for general acid-base ...
|
|
5
|
Do enzymes change the nature of transition states? Mapping the transition state for general acid-base ...
|
|
6
|
Integrin alphaMbeta2 orchestrates and accelerates plasminogen activation and fibrinolysis by neutrophils
|
|
7
|
Integrin alphaMbeta2 orchestrates and accelerates plasminogen activation and fibrinolysis by neutrophils
|
|
8
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
9
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
10
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
11
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
12
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
13
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
14
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
15
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
16
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
17
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
18
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
19
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
20
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
21
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
22
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
23
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
24
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
25
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
26
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
27
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
28
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
29
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
30
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
31
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
32
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
33
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
34
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
35
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
36
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
37
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
38
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
39
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
40
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
41
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
42
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
43
|
Role of glycine 81 in (S)-mandelate dehydrogenase from Pseudomonas putida in substrate specificity and oxidase ...
|
|
44
|
Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from ...
|
|
45
|
Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from ...
|
|
46
|
Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from ...
|
|
47
|
Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from ...
|
|
48
|
Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from ...
|
|
49
|
Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from ...
|
|
50
|
Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from ...
|
|
51
|
Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from ...
|
|
52
|
Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from ...
|
|
53
|
On the relationship between affinity for molecular hydrogen and the physiological directionality of ...
|
|
54
|
On the relationship between affinity for molecular hydrogen and the physiological directionality of ...
|
|
55
|
Identification of arginine residues important for the activity of Escherichia coli signal peptidase I
|
|
56
|
Identification of arginine residues important for the activity of Escherichia coli signal peptidase I
|
|
57
|
Identification of arginine residues important for the activity of Escherichia coli signal peptidase I
|
|
58
|
Identification of arginine residues important for the activity of Escherichia coli signal peptidase I
|
|
59
|
Identification of arginine residues important for the activity of Escherichia coli signal peptidase I
|
|
60
|
Identification of arginine residues important for the activity of Escherichia coli signal peptidase I
|
|
61
|
Identification of arginine residues important for the activity of Escherichia coli signal peptidase I
|
|
62
|
Identification of arginine residues important for the activity of Escherichia coli signal peptidase I
|
|
63
|
Identification of arginine residues important for the activity of Escherichia coli signal peptidase I
|
|
64
|
Identification of arginine residues important for the activity of Escherichia coli signal peptidase I
|
|
65
|
Identification of arginine residues important for the activity of Escherichia coli signal peptidase I
|
|
66
|
Identification of arginine residues important for the activity of Escherichia coli signal peptidase I
|
|
67
|
Identification of arginine residues important for the activity of Escherichia coli signal peptidase I
|
|
68
|
Identification of arginine residues important for the activity of Escherichia coli signal peptidase I
|
|
69
|
Identification of arginine residues important for the activity of Escherichia coli signal peptidase I
|
|
70
|
Identification of arginine residues important for the activity of Escherichia coli signal peptidase I
|
|
71
|
Identification of arginine residues important for the activity of Escherichia coli signal peptidase I
|
|
72
|
Identification of arginine residues important for the activity of Escherichia coli signal peptidase I
|
|
73
|
Protein engineering of pyruvate carboxylase: investigation on the function of acetyl-CoA and the quaternary ...
|
|
74
|
Protein engineering of pyruvate carboxylase: investigation on the function of acetyl-CoA and the quaternary ...
|
|
75
|
Protein engineering of pyruvate carboxylase: investigation on the function of acetyl-CoA and the quaternary ...
|
|
76
|
Protein engineering of pyruvate carboxylase: investigation on the function of acetyl-CoA and the quaternary ...
|
|
77
|
Protein engineering of pyruvate carboxylase: investigation on the function of acetyl-CoA and the quaternary ...
|
|
78
|
Protein engineering of pyruvate carboxylase: investigation on the function of acetyl-CoA and the quaternary ...
|
|
79
|
Protein engineering of pyruvate carboxylase: investigation on the function of acetyl-CoA and the quaternary ...
|
|
80
|
Protein engineering of pyruvate carboxylase: investigation on the function of acetyl-CoA and the quaternary ...
|
|
81
|
The mechanism of action of the fragile histidine triad, Fhit: isolation of a covalent adenylyl enzyme and ...
|
|
82
|
The mechanism of action of the fragile histidine triad, Fhit: isolation of a covalent adenylyl enzyme and ...
|
|
83
|
The mechanism of action of the fragile histidine triad, Fhit: isolation of a covalent adenylyl enzyme and ...
|
|
84
|
The mechanism of action of the fragile histidine triad, Fhit: isolation of a covalent adenylyl enzyme and ...
|
|
85
|
The mechanism of action of the fragile histidine triad, Fhit: isolation of a covalent adenylyl enzyme and ...
|
|
86
|
The mechanism of action of the fragile histidine triad, Fhit: isolation of a covalent adenylyl enzyme and ...
|
|
87
|
Reaction mechanism of chitobiose phosphorylase from Vibrio proteolyticus: identification of family 36 ...
|
|
88
|
Reaction mechanism of chitobiose phosphorylase from Vibrio proteolyticus: identification of family 36 ...
|
|
89
|
Reaction mechanism of chitobiose phosphorylase from Vibrio proteolyticus: identification of family 36 ...
|
|
90
|
Reaction mechanism of chitobiose phosphorylase from Vibrio proteolyticus: identification of family 36 ...
|
|
91
|
Reaction mechanism of chitobiose phosphorylase from Vibrio proteolyticus: identification of family 36 ...
|
|
92
|
Reaction mechanism of chitobiose phosphorylase from Vibrio proteolyticus: identification of family 36 ...
|
|
93
|
Defining the binding site of homotetrameric R67 dihydrofolate reductase and correlating binding enthalpy with ...
|
|
94
|
Defining the binding site of homotetrameric R67 dihydrofolate reductase and correlating binding enthalpy with ...
|
|
95
|
Defining the binding site of homotetrameric R67 dihydrofolate reductase and correlating binding enthalpy with ...
|
|
96
|
Defining the binding site of homotetrameric R67 dihydrofolate reductase and correlating binding enthalpy with ...
|
|
97
|
Defining the binding site of homotetrameric R67 dihydrofolate reductase and correlating binding enthalpy with ...
|
|
98
|
Defining the binding site of homotetrameric R67 dihydrofolate reductase and correlating binding enthalpy with ...
|
|
99
|
Defining the binding site of homotetrameric R67 dihydrofolate reductase and correlating binding enthalpy with ...
|
|
100
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
101
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
102
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
103
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
104
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
105
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
106
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
107
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
108
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
109
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
110
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
111
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
112
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
113
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
114
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
115
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
116
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
117
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
118
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
119
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
120
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
121
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
122
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
123
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
124
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
125
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
126
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
127
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
128
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
129
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
130
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
131
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
132
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
133
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
134
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
135
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
136
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
137
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
138
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
139
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
140
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
141
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
142
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
143
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
144
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
145
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
146
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
147
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
148
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
149
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
150
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
151
|
Probing the interactions between the folding elements early in the folding of Escherichia coli dihydrofolate ...
|
|
152
|
Kinetic evidence related to substrate-assisted catalysis of family 18 chitinases
|
|
153
|
Kinetic evidence related to substrate-assisted catalysis of family 18 chitinases
|
|
154
|
Kinetic evidence related to substrate-assisted catalysis of family 18 chitinases
|
|
155
|
Changing the metal ion selectivity of rabbit muscle enolase by mutagenesis: effects of zhe G37A and G41A ...
|
|
156
|
Changing the metal ion selectivity of rabbit muscle enolase by mutagenesis: effects of zhe G37A and G41A ...
|
|
157
|
Changing the metal ion selectivity of rabbit muscle enolase by mutagenesis: effects of zhe G37A and G41A ...
|
|
158
|
Changing the metal ion selectivity of rabbit muscle enolase by mutagenesis: effects of zhe G37A and G41A ...
|
|
159
|
Changing the metal ion selectivity of rabbit muscle enolase by mutagenesis: effects of zhe G37A and G41A ...
|
|
160
|
Changing the metal ion selectivity of rabbit muscle enolase by mutagenesis: effects of zhe G37A and G41A ...
|
|
161
|
Changing the metal ion selectivity of rabbit muscle enolase by mutagenesis: effects of zhe G37A and G41A ...
|
|
162
|
Changing the metal ion selectivity of rabbit muscle enolase by mutagenesis: effects of zhe G37A and G41A ...
|
|
163
|
Changing the metal ion selectivity of rabbit muscle enolase by mutagenesis: effects of zhe G37A and G41A ...
|
|
164
|
Changing the metal ion selectivity of rabbit muscle enolase by mutagenesis: effects of zhe G37A and G41A ...
|
|
165
|
Changing the metal ion selectivity of rabbit muscle enolase by mutagenesis: effects of zhe G37A and G41A ...
|
|
166
|
Changing the metal ion selectivity of rabbit muscle enolase by mutagenesis: effects of zhe G37A and G41A ...
|
|
167
|
Changing the metal ion selectivity of rabbit muscle enolase by mutagenesis: effects of zhe G37A and G41A ...
|
|
168
|
Changing the metal ion selectivity of rabbit muscle enolase by mutagenesis: effects of zhe G37A and G41A ...
|
|
169
|
Changing the metal ion selectivity of rabbit muscle enolase by mutagenesis: effects of zhe G37A and G41A ...
|
|
172
|
Characterization of Mycobacterium tuberculosis NAD kinase: functional analysis of the full-length enzyme by ...
|
|
173
|
Characterization of Mycobacterium tuberculosis NAD kinase: functional analysis of the full-length enzyme by ...
|
|
174
|
Characterization of Mycobacterium tuberculosis NAD kinase: functional analysis of the full-length enzyme by ...
|
|
175
|
Characterization of Mycobacterium tuberculosis NAD kinase: functional analysis of the full-length enzyme by ...
|
|
176
|
Characterization of Mycobacterium tuberculosis NAD kinase: functional analysis of the full-length enzyme by ...
|
|
177
|
Characterization of Mycobacterium tuberculosis NAD kinase: functional analysis of the full-length enzyme by ...
|
|
178
|
Characterization of Mycobacterium tuberculosis NAD kinase: functional analysis of the full-length enzyme by ...
|
|
179
|
Characterization of Mycobacterium tuberculosis NAD kinase: functional analysis of the full-length enzyme by ...
|
|
180
|
Characterization of Mycobacterium tuberculosis NAD kinase: functional analysis of the full-length enzyme by ...
|
|
181
|
Characterization of Mycobacterium tuberculosis NAD kinase: functional analysis of the full-length enzyme by ...
|
|
182
|
Characterization of Mycobacterium tuberculosis NAD kinase: functional analysis of the full-length enzyme by ...
|
|
183
|
Characterization of Mycobacterium tuberculosis NAD kinase: functional analysis of the full-length enzyme by ...
|
|
184
|
Characterization of Mycobacterium tuberculosis NAD kinase: functional analysis of the full-length enzyme by ...
|
|
185
|
Characterization of Mycobacterium tuberculosis NAD kinase: functional analysis of the full-length enzyme by ...
|
|
186
|
Characterization of Mycobacterium tuberculosis NAD kinase: functional analysis of the full-length enzyme by ...
|
|
187
|
Characterization of Mycobacterium tuberculosis NAD kinase: functional analysis of the full-length enzyme by ...
|
|
188
|
Kinetics and comparative reactivity of human class I and class IIb histone deacetylases
|
|
189
|
Kinetics and comparative reactivity of human class I and class IIb histone deacetylases
|
|
190
|
Kinetics and comparative reactivity of human class I and class IIb histone deacetylases
|
|
191
|
Kinetics and comparative reactivity of human class I and class IIb histone deacetylases
|
|
192
|
Kinetics and comparative reactivity of human class I and class IIb histone deacetylases
|
|
193
|
Kinetics and comparative reactivity of human class I and class IIb histone deacetylases
|
|
194
|
Structural and mechanistic studies of chloride induced activation of human pancreatic alpha-amylase
|
|
195
|
Structural and mechanistic studies of chloride induced activation of human pancreatic alpha-amylase
|
|
196
|
Effects of AMP and fructose 2,6-bisphosphate on fluxes between glucose 6-phosphate and triose-phosphate in ...
|
|
197
|
Effects of AMP and fructose 2,6-bisphosphate on fluxes between glucose 6-phosphate and triose-phosphate in ...
|
|
198
|
Effects of AMP and fructose 2,6-bisphosphate on fluxes between glucose 6-phosphate and triose-phosphate in ...
|
|
199
|
Effects of AMP and fructose 2,6-bisphosphate on fluxes between glucose 6-phosphate and triose-phosphate in ...
|
|
200
|
Effects of AMP and fructose 2,6-bisphosphate on fluxes between glucose 6-phosphate and triose-phosphate in ...
|
|
201
|
Effects of AMP and fructose 2,6-bisphosphate on fluxes between glucose 6-phosphate and triose-phosphate in ...
|
|
202
|
Effects of AMP and fructose 2,6-bisphosphate on fluxes between glucose 6-phosphate and triose-phosphate in ...
|
|
203
|
Effects of AMP and fructose 2,6-bisphosphate on fluxes between glucose 6-phosphate and triose-phosphate in ...
|
|
204
|
Effects of AMP and fructose 2,6-bisphosphate on fluxes between glucose 6-phosphate and triose-phosphate in ...
|
|
205
|
Effects of AMP and fructose 2,6-bisphosphate on fluxes between glucose 6-phosphate and triose-phosphate in ...
|
|
206
|
Effects of AMP and fructose 2,6-bisphosphate on fluxes between glucose 6-phosphate and triose-phosphate in ...
|
|
207
|
Effects of AMP and fructose 2,6-bisphosphate on fluxes between glucose 6-phosphate and triose-phosphate in ...
|
|
208
|
Effects of AMP and fructose 2,6-bisphosphate on fluxes between glucose 6-phosphate and triose-phosphate in ...
|
|
209
|
Effects of AMP and fructose 2,6-bisphosphate on fluxes between glucose 6-phosphate and triose-phosphate in ...
|
|
210
|
Effects of AMP and fructose 2,6-bisphosphate on fluxes between glucose 6-phosphate and triose-phosphate in ...
|
|
211
|
Effects of AMP and fructose 2,6-bisphosphate on fluxes between glucose 6-phosphate and triose-phosphate in ...
|
|
212
|
Rates of elementary catalytic steps for different metal forms of the family II pyrophosphatase from ...
|
|
213
|
Rates of elementary catalytic steps for different metal forms of the family II pyrophosphatase from ...
|
|
214
|
Rates of elementary catalytic steps for different metal forms of the family II pyrophosphatase from ...
|
|
215
|
Rates of elementary catalytic steps for different metal forms of the family II pyrophosphatase from ...
|
|
216
|
Rates of elementary catalytic steps for different metal forms of the family II pyrophosphatase from ...
|
|
217
|
Rates of elementary catalytic steps for different metal forms of the family II pyrophosphatase from ...
|
|
218
|
Rates of elementary catalytic steps for different metal forms of the family II pyrophosphatase from ...
|
|
219
|
Rates of elementary catalytic steps for different metal forms of the family II pyrophosphatase from ...
|
|
220
|
Rates of elementary catalytic steps for different metal forms of the family II pyrophosphatase from ...
|
|
221
|
Rates of elementary catalytic steps for different metal forms of the family II pyrophosphatase from ...
|
|
222
|
Rates of elementary catalytic steps for different metal forms of the family II pyrophosphatase from ...
|
|
223
|
Rates of elementary catalytic steps for different metal forms of the family II pyrophosphatase from ...
|
|
224
|
Rates of elementary catalytic steps for different metal forms of the family II pyrophosphatase from ...
|
|
225
|
Peroxidase catalyzed nitration of tryptophan derivatives. Mechanism, products and comparison with chemical ...
|
|
226
|
Peroxidase catalyzed nitration of tryptophan derivatives. Mechanism, products and comparison with chemical ...
|
|
227
|
Peroxidase catalyzed nitration of tryptophan derivatives. Mechanism, products and comparison with chemical ...
|
|
228
|
Peroxidase catalyzed nitration of tryptophan derivatives. Mechanism, products and comparison with chemical ...
|
|
229
|
Peroxidase catalyzed nitration of tryptophan derivatives. Mechanism, products and comparison with chemical ...
|
|
230
|
Peroxidase catalyzed nitration of tryptophan derivatives. Mechanism, products and comparison with chemical ...
|
|
231
|
Peroxidase catalyzed nitration of tryptophan derivatives. Mechanism, products and comparison with chemical ...
|
|
232
|
Peroxidase catalyzed nitration of tryptophan derivatives. Mechanism, products and comparison with chemical ...
|
|
233
|
Peroxidase catalyzed nitration of tryptophan derivatives. Mechanism, products and comparison with chemical ...
|
|
234
|
Peroxidase catalyzed nitration of tryptophan derivatives. Mechanism, products and comparison with chemical ...
|
|
235
|
Peroxidase catalyzed nitration of tryptophan derivatives. Mechanism, products and comparison with chemical ...
|
|
236
|
Peroxidase catalyzed nitration of tryptophan derivatives. Mechanism, products and comparison with chemical ...
|
|
237
|
Catalytic and structural contributions for glutathione-binding residues in a Delta class glutathione ...
|
|
238
|
Catalytic and structural contributions for glutathione-binding residues in a Delta class glutathione ...
|
|
239
|
Catalytic and structural contributions for glutathione-binding residues in a Delta class glutathione ...
|
|
240
|
Catalytic and structural contributions for glutathione-binding residues in a Delta class glutathione ...
|
|
241
|
Catalytic and structural contributions for glutathione-binding residues in a Delta class glutathione ...
|
|
242
|
Catalytic and structural contributions for glutathione-binding residues in a Delta class glutathione ...
|
|
243
|
Catalytic and structural contributions for glutathione-binding residues in a Delta class glutathione ...
|
|
244
|
Catalytic and structural contributions for glutathione-binding residues in a Delta class glutathione ...
|
|
245
|
Catalytic and structural contributions for glutathione-binding residues in a Delta class glutathione ...
|
|
246
|
Catalytic and structural contributions for glutathione-binding residues in a Delta class glutathione ...
|
|
247
|
A novel beta-lactamase activity from a penicillin-binding protein of Treponema pallidum and why syphilis is ...
|
|
248
|
A novel beta-lactamase activity from a penicillin-binding protein of Treponema pallidum and why syphilis is ...
|
|
249
|
A novel beta-lactamase activity from a penicillin-binding protein of Treponema pallidum and why syphilis is ...
|
|
250
|
A novel beta-lactamase activity from a penicillin-binding protein of Treponema pallidum and why syphilis is ...
|
|
251
|
A novel beta-lactamase activity from a penicillin-binding protein of Treponema pallidum and why syphilis is ...
|
|
252
|
A novel beta-lactamase activity from a penicillin-binding protein of Treponema pallidum and why syphilis is ...
|
|
253
|
A novel beta-lactamase activity from a penicillin-binding protein of Treponema pallidum and why syphilis is ...
|
|
254
|
A novel beta-lactamase activity from a penicillin-binding protein of Treponema pallidum and why syphilis is ...
|
|
255
|
A novel beta-lactamase activity from a penicillin-binding protein of Treponema pallidum and why syphilis is ...
|
|
256
|
A novel beta-lactamase activity from a penicillin-binding protein of Treponema pallidum and why syphilis is ...
|
|
257
|
A novel beta-lactamase activity from a penicillin-binding protein of Treponema pallidum and why syphilis is ...
|
|
258
|
A novel beta-lactamase activity from a penicillin-binding protein of Treponema pallidum and why syphilis is ...
|
|
259
|
Minimization of cavity size ensures protein stability and folding: structures of Phe46-replaced bovine ...
|
|
260
|
Minimization of cavity size ensures protein stability and folding: structures of Phe46-replaced bovine ...
|
|
261
|
Minimization of cavity size ensures protein stability and folding: structures of Phe46-replaced bovine ...
|
|
262
|
Minimization of cavity size ensures protein stability and folding: structures of Phe46-replaced bovine ...
|
|
263
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
264
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
265
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
266
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
267
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
268
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
269
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
270
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
271
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
272
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
273
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
274
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
275
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
276
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
277
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
278
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
279
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
280
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
281
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
282
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
283
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
284
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
285
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
286
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
287
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
288
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
289
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
290
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
291
|
Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha ...
|
|
292
|
Biochemical characterization of WbpA, a UDP-N-acetyl-D-glucosamine 6-dehydrogenase involved in O-antigen ...
|
|
293
|
Biochemical characterization of WbpA, a UDP-N-acetyl-D-glucosamine 6-dehydrogenase involved in O-antigen ...
|
|
294
|
Kinetic characterization of yeast pyruvate carboxylase isozyme Pyc1 and the Pyc1 mutant, C249A
|
|
295
|
Kinetic characterization of yeast pyruvate carboxylase isozyme Pyc1 and the Pyc1 mutant, C249A
|
|
296
|
Kinetic characterization of yeast pyruvate carboxylase isozyme Pyc1 and the Pyc1 mutant, C249A
|
|
297
|
Kinetic characterization of yeast pyruvate carboxylase isozyme Pyc1 and the Pyc1 mutant, C249A
|
|
298
|
Kinetic characterization of yeast pyruvate carboxylase isozyme Pyc1 and the Pyc1 mutant, C249A
|
|
299
|
Kinetic characterization of yeast pyruvate carboxylase isozyme Pyc1 and the Pyc1 mutant, C249A
|
|
300
|
Kinetic characterization of yeast pyruvate carboxylase isozyme Pyc1 and the Pyc1 mutant, C249A
|
|
301
|
Kinetic characterization of yeast pyruvate carboxylase isozyme Pyc1 and the Pyc1 mutant, C249A
|
|
302
|
Kinetic characterization of yeast pyruvate carboxylase isozyme Pyc1 and the Pyc1 mutant, C249A
|
|
303
|
Kinetic characterization of yeast pyruvate carboxylase isozyme Pyc1 and the Pyc1 mutant, C249A
|
|
304
|
Dihydrodipicolinate synthase (DHDPS) from Escherichia coli displays partial mixed inhibition with respect to ...
|
|
305
|
Dihydrodipicolinate synthase (DHDPS) from Escherichia coli displays partial mixed inhibition with respect to ...
|
|
306
|
Dihydrodipicolinate synthase (DHDPS) from Escherichia coli displays partial mixed inhibition with respect to ...
|
|
307
|
Different properties of the central and peripheral forms of human tryptophan hydroxylase
|
|
308
|
Different properties of the central and peripheral forms of human tryptophan hydroxylase
|
|
309
|
Different properties of the central and peripheral forms of human tryptophan hydroxylase
|
|
310
|
Different properties of the central and peripheral forms of human tryptophan hydroxylase
|
|
311
|
Different properties of the central and peripheral forms of human tryptophan hydroxylase
|
|
312
|
Different properties of the central and peripheral forms of human tryptophan hydroxylase
|
|
313
|
Different properties of the central and peripheral forms of human tryptophan hydroxylase
|
|
314
|
Different properties of the central and peripheral forms of human tryptophan hydroxylase
|
|
315
|
Different properties of the central and peripheral forms of human tryptophan hydroxylase
|
|
316
|
Different properties of the central and peripheral forms of human tryptophan hydroxylase
|
|
317
|
Different properties of the central and peripheral forms of human tryptophan hydroxylase
|
|
318
|
Different properties of the central and peripheral forms of human tryptophan hydroxylase
|
|
319
|
Different properties of the central and peripheral forms of human tryptophan hydroxylase
|
|
320
|
Different properties of the central and peripheral forms of human tryptophan hydroxylase
|
|
321
|
Different properties of the central and peripheral forms of human tryptophan hydroxylase
|
|
322
|
Different properties of the central and peripheral forms of human tryptophan hydroxylase
|
|
323
|
Different properties of the central and peripheral forms of human tryptophan hydroxylase
|
|
324
|
Different properties of the central and peripheral forms of human tryptophan hydroxylase
|
|
325
|
A metallo-beta-lactamase enzyme in action: crystal structures of the monozinc carbapenemase CphA and its ...
|
|
326
|
A metallo-beta-lactamase enzyme in action: crystal structures of the monozinc carbapenemase CphA and its ...
|
|
327
|
A metallo-beta-lactamase enzyme in action: crystal structures of the monozinc carbapenemase CphA and its ...
|
|
328
|
A metallo-beta-lactamase enzyme in action: crystal structures of the monozinc carbapenemase CphA and its ...
|
|
329
|
Trypanothione Synthesis in Crithidia Revisited
|
|
330
|
Trypanothione Synthesis in Crithidia Revisited
|
|
407
|
Characterization of a Novel NADP+- Dependent D-Arabitol Dehydrogenase from the Plant Pathogen Uromyces fabae
|
|
408
|
Characterization of a Novel NADP+- Dependent D-Arabitol Dehydrogenase from the Plant Pathogen Uromyces fabae
|
|
409
|
Characterization of a Novel NADP+- Dependent D-Arabitol Dehydrogenase from the Plant Pathogen Uromyces fabae
|
|
410
|
Characterization of a Novel NADP+- Dependent D-Arabitol Dehydrogenase from the Plant Pathogen Uromyces fabae
|
|
411
|
Characterization of a Novel NADP+- Dependent D-Arabitol Dehydrogenase from the Plant Pathogen Uromyces fabae
|
|
412
|
Characterization of a Novel NADP+- Dependent D-Arabitol Dehydrogenase from the Plant Pathogen Uromyces fabae
|
|
413
|
The role of active site glutamate residues in catalysis of Rhodobacter capsulatus xanthine dehydrogenase
|
|
414
|
The role of active site glutamate residues in catalysis of Rhodobacter capsulatus xanthine dehydrogenase
|
|
415
|
The role of active site glutamate residues in catalysis of Rhodobacter capsulatus xanthine dehydrogenase
|
|
416
|
The role of active site glutamate residues in catalysis of Rhodobacter capsulatus xanthine dehydrogenase
|
|
428
|
Kinetic Studies of DNA Cleavage Reactions Catalyzed by an ATP-Dependent Deoxyribonuclease on a 27-MHz ...
|
|
429
|
Kinetic Studies of DNA Cleavage Reactions Catalyzed by an ATP-Dependent Deoxyribonuclease on a 27-MHz ...
|
|
430
|
Kinetic Studies of DNA Cleavage Reactions Catalyzed by an ATP-Dependent Deoxyribonuclease on a 27-MHz ...
|
|
431
|
Kinetic Studies of DNA Cleavage Reactions Catalyzed by an ATP-Dependent Deoxyribonuclease on a 27-MHz ...
|
|
432
|
Kinetic Studies of DNA Cleavage Reactions Catalyzed by an ATP-Dependent Deoxyribonuclease on a 27-MHz ...
|
|
433
|
Kinetic Studies of DNA Cleavage Reactions Catalyzed by an ATP-Dependent Deoxyribonuclease on a 27-MHz ...
|
|
434
|
Human Carbonyl Reductase Catalyzes Reduction of 4-Oxonon-2-enal
|
|
435
|
Human Carbonyl Reductase Catalyzes Reduction of 4-Oxonon-2-enal
|
|
436
|
Human Carbonyl Reductase Catalyzes Reduction of 4-Oxonon-2-enal
|
|
437
|
Human Carbonyl Reductase Catalyzes Reduction of 4-Oxonon-2-enal
|
|
438
|
Human Carbonyl Reductase Catalyzes Reduction of 4-Oxonon-2-enal
|
|
439
|
Human Carbonyl Reductase Catalyzes Reduction of 4-Oxonon-2-enal
|
|
443
|
A novel assay method for an amino acid racemase based on circular dichroism
|
|
444
|
A novel assay method for an amino acid racemase based on circular dichroism
|
|
445
|
A novel assay method for an amino acid racemase based on circular dichroism
|
|
446
|
Human acetyl-coenzyme A:alpha-glucosaminide N-acetyltransferase. Kinetic characterization and mechanistic ...
|
|
447
|
Human acetyl-coenzyme A:alpha-glucosaminide N-acetyltransferase. Kinetic characterization and mechanistic ...
|
|
456
|
Role of Methionine-13 in the Catalytic Mechanism of 6-Phosphogluconate Dehydrogenase from Sheep Liver
|
|
457
|
Role of Methionine-13 in the Catalytic Mechanism of 6-Phosphogluconate Dehydrogenase from Sheep Liver
|
|
458
|
Role of Methionine-13 in the Catalytic Mechanism of 6-Phosphogluconate Dehydrogenase from Sheep Liver
|
|
459
|
Role of Methionine-13 in the Catalytic Mechanism of 6-Phosphogluconate Dehydrogenase from Sheep Liver
|
|
460
|
Role of Methionine-13 in the Catalytic Mechanism of 6-Phosphogluconate Dehydrogenase from Sheep Liver
|
|
461
|
Role of Methionine-13 in the Catalytic Mechanism of 6-Phosphogluconate Dehydrogenase from Sheep Liver
|
|
474
|
Dissection of the Steps of Phospholipase C beta 2 Activity That Are Enhanced by G beta gamma Subunits
|
|
475
|
Dissection of the Steps of Phospholipase C beta 2 Activity That Are Enhanced by G beta gamma Subunits
|
|
476
|
Dissection of the Steps of Phospholipase C beta 2 Activity That Are Enhanced by G beta gamma Subunits
|
|
477
|
Dissection of the Steps of Phospholipase C beta 2 Activity That Are Enhanced by G beta gamma Subunits
|
|
478
|
Dissection of the Steps of Phospholipase C beta 2 Activity That Are Enhanced by G beta gamma Subunits
|
|
479
|
Effect of cadmium on ferredoxin:NADP+ oxidoreductase activity
|
|
480
|
Effect of cadmium on ferredoxin:NADP+ oxidoreductase activity
|
|
481
|
Effect of cadmium on ferredoxin:NADP+ oxidoreductase activity
|
|
482
|
Effect of cadmium on ferredoxin:NADP+ oxidoreductase activity
|
|
483
|
Mechanistic and structural analysis of a family 31 alpha-glycosidase and its glycosyl-enzyme intermediate
|
|
484
|
Mechanistic and structural analysis of a family 31 alpha-glycosidase and its glycosyl-enzyme intermediate
|
|
485
|
Mechanistic and structural analysis of a family 31 alpha-glycosidase and its glycosyl-enzyme intermediate
|
|
486
|
Characterization of N-acetylneuraminic acid synthase isoenzyme 1 from Campylobacter jejuni
|
|
487
|
Characterization of N-acetylneuraminic acid synthase isoenzyme 1 from Campylobacter jejuni
|
|
488
|
Characterization of N-acetylneuraminic acid synthase isoenzyme 1 from Campylobacter jejuni
|
|
489
|
Characterization of N-acetylneuraminic acid synthase isoenzyme 1 from Campylobacter jejuni
|
|
490
|
Biophysical and kinetic analysis of wild-type and site- directed mutants of the isolated and native ...
|
|
491
|
Biophysical and kinetic analysis of wild-type and site- directed mutants of the isolated and native ...
|
|
492
|
Biophysical and kinetic analysis of wild-type and site- directed mutants of the isolated and native ...
|
|
493
|
Biophysical and kinetic analysis of wild-type and site- directed mutants of the isolated and native ...
|
|
494
|
Biophysical and kinetic analysis of wild-type and site- directed mutants of the isolated and native ...
|
|
495
|
Biophysical and kinetic analysis of wild-type and site- directed mutants of the isolated and native ...
|
|
496
|
Mechanistic differences in inhibition of ubiquinol cytochrome c reductase by the proximal Qo-site inhibitors ...
|
|
497
|
Mechanistic differences in inhibition of ubiquinol cytochrome c reductase by the proximal Qo-site inhibitors ...
|
|
498
|
Mechanistic differences in inhibition of ubiquinol cytochrome c reductase by the proximal Qo-site inhibitors ...
|
|
499
|
L-Pipecolic acid metabolism in human liver: L-alpha-aminoadipate delta-semialdehyde oxidoreductase
|
|
500
|
L-Pipecolic acid metabolism in human liver: L-alpha-aminoadipate delta-semialdehyde oxidoreductase
|
|
513
|
Use of 8-substituted-FAD analogues to investigate the hydroxylation mechanism of the flavoprotein ...
|
|
514
|
Use of 8-substituted-FAD analogues to investigate the hydroxylation mechanism of the flavoprotein ...
|
|
515
|
Use of 8-substituted-FAD analogues to investigate the hydroxylation mechanism of the flavoprotein ...
|
|
516
|
Use of 8-substituted-FAD analogues to investigate the hydroxylation mechanism of the flavoprotein ...
|
|
517
|
Use of 8-substituted-FAD analogues to investigate the hydroxylation mechanism of the flavoprotein ...
|
|
518
|
Use of 8-substituted-FAD analogues to investigate the hydroxylation mechanism of the flavoprotein ...
|
|
519
|
Use of 8-substituted-FAD analogues to investigate the hydroxylation mechanism of the flavoprotein ...
|
|
520
|
Use of 8-substituted-FAD analogues to investigate the hydroxylation mechanism of the flavoprotein ...
|
|
521
|
Use of 8-substituted-FAD analogues to investigate the hydroxylation mechanism of the flavoprotein ...
|
|
522
|
Use of 8-substituted-FAD analogues to investigate the hydroxylation mechanism of the flavoprotein ...
|
|
523
|
Use of 8-substituted-FAD analogues to investigate the hydroxylation mechanism of the flavoprotein ...
|
|
524
|
Use of 8-substituted-FAD analogues to investigate the hydroxylation mechanism of the flavoprotein ...
|
|
525
|
Use of 8-substituted-FAD analogues to investigate the hydroxylation mechanism of the flavoprotein ...
|
|
526
|
Use of 8-substituted-FAD analogues to investigate the hydroxylation mechanism of the flavoprotein ...
|
|
527
|
Use of 8-substituted-FAD analogues to investigate the hydroxylation mechanism of the flavoprotein ...
|
|
528
|
Amine-synthesizing enzyme N-substituted formamide deformylase: screening, purification, characterization, and ...
|
|
529
|
Amine-synthesizing enzyme N-substituted formamide deformylase: screening, purification, characterization, and ...
|
|
530
|
Mutational analysis of peptidyl carrier protein and acyl carrier protein synthase unveils residues involved in ...
|
|
531
|
Mutational analysis of peptidyl carrier protein and acyl carrier protein synthase unveils residues involved in ...
|
|
532
|
Mutational analysis of peptidyl carrier protein and acyl carrier protein synthase unveils residues involved in ...
|
|
533
|
Mutational analysis of peptidyl carrier protein and acyl carrier protein synthase unveils residues involved in ...
|
|
534
|
Mutational analysis of peptidyl carrier protein and acyl carrier protein synthase unveils residues involved in ...
|
|
535
|
Mutational analysis of peptidyl carrier protein and acyl carrier protein synthase unveils residues involved in ...
|
|
536
|
Mutational analysis of peptidyl carrier protein and acyl carrier protein synthase unveils residues involved in ...
|
|
537
|
Mutational analysis of peptidyl carrier protein and acyl carrier protein synthase unveils residues involved in ...
|
|
538
|
Mutational analysis of peptidyl carrier protein and acyl carrier protein synthase unveils residues involved in ...
|
|
539
|
Mutational analysis of peptidyl carrier protein and acyl carrier protein synthase unveils residues involved in ...
|
|
540
|
Mutational analysis of peptidyl carrier protein and acyl carrier protein synthase unveils residues involved in ...
|
|
541
|
Mutational analysis of peptidyl carrier protein and acyl carrier protein synthase unveils residues involved in ...
|
|
542
|
Mutational analysis of peptidyl carrier protein and acyl carrier protein synthase unveils residues involved in ...
|
|
543
|
Mutational analysis of peptidyl carrier protein and acyl carrier protein synthase unveils residues involved in ...
|
|
544
|
Mutational analysis of peptidyl carrier protein and acyl carrier protein synthase unveils residues involved in ...
|
|
545
|
Mutational analysis of peptidyl carrier protein and acyl carrier protein synthase unveils residues involved in ...
|
|
546
|
Mutational analysis of peptidyl carrier protein and acyl carrier protein synthase unveils residues involved in ...
|
|
547
|
Mutational analysis of peptidyl carrier protein and acyl carrier protein synthase unveils residues involved in ...
|
|
548
|
Mutational analysis of peptidyl carrier protein and acyl carrier protein synthase unveils residues involved in ...
|
|
557
|
Hexose-6-phosphate dehydrogenase determines the reaction direction of 11beta-hydroxysteroid dehydrogenase type ...
|
|
558
|
Hexose-6-phosphate dehydrogenase determines the reaction direction of 11beta-hydroxysteroid dehydrogenase type ...
|
|
559
|
Hexose-6-phosphate dehydrogenase determines the reaction direction of 11beta-hydroxysteroid dehydrogenase type ...
|
|
560
|
Hexose-6-phosphate dehydrogenase determines the reaction direction of 11beta-hydroxysteroid dehydrogenase type ...
|
|
561
|
Hexose-6-phosphate dehydrogenase determines the reaction direction of 11beta-hydroxysteroid dehydrogenase type ...
|
|
562
|
Hexose-6-phosphate dehydrogenase determines the reaction direction of 11beta-hydroxysteroid dehydrogenase type ...
|
|
563
|
Hexose-6-phosphate dehydrogenase determines the reaction direction of 11beta-hydroxysteroid dehydrogenase type ...
|
|
564
|
Hexose-6-phosphate dehydrogenase determines the reaction direction of 11beta-hydroxysteroid dehydrogenase type ...
|
|
565
|
Hexose-6-phosphate dehydrogenase determines the reaction direction of 11beta-hydroxysteroid dehydrogenase type ...
|
|
566
|
Hexose-6-phosphate dehydrogenase determines the reaction direction of 11beta-hydroxysteroid dehydrogenase type ...
|
|
567
|
Hexose-6-phosphate dehydrogenase determines the reaction direction of 11beta-hydroxysteroid dehydrogenase type ...
|
|
568
|
Hexose-6-phosphate dehydrogenase determines the reaction direction of 11beta-hydroxysteroid dehydrogenase type ...
|
|
590
|
Enterococcus faecalis mevalonate kinase
|
|
615
|
Rapid evolution of beta-glucuronidase specificity by saturation mutagenesis of an active site loop
|
|
616
|
Rapid evolution of beta-glucuronidase specificity by saturation mutagenesis of an active site loop
|
|
617
|
Rapid evolution of beta-glucuronidase specificity by saturation mutagenesis of an active site loop
|
|
618
|
Rapid evolution of beta-glucuronidase specificity by saturation mutagenesis of an active site loop
|
|
619
|
Rapid evolution of beta-glucuronidase specificity by saturation mutagenesis of an active site loop
|
|
620
|
Rapid evolution of beta-glucuronidase specificity by saturation mutagenesis of an active site loop
|
|
621
|
Rapid evolution of beta-glucuronidase specificity by saturation mutagenesis of an active site loop
|
|
622
|
Rapid evolution of beta-glucuronidase specificity by saturation mutagenesis of an active site loop
|
|
623
|
Rapid evolution of beta-glucuronidase specificity by saturation mutagenesis of an active site loop
|
|
624
|
Rapid evolution of beta-glucuronidase specificity by saturation mutagenesis of an active site loop
|
|
625
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
626
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
627
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
628
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
629
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
630
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
631
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
632
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
633
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
634
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
635
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
636
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
637
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
638
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
639
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
640
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
641
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
642
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
643
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
644
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
645
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
646
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
647
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
648
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
649
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
650
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
651
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
652
|
Probing the dynamics of a mobile loop above the active site of L1, a metallo-beta-lactamase from ...
|
|
653
|
UDP-glucuronic Acid:Anthocyanin Glucuronosyltransferase from Red Daisy (Bellis perennis) Flowers
|
|
654
|
UDP-glucuronic Acid:Anthocyanin Glucuronosyltransferase from Red Daisy (Bellis perennis) Flowers
|
|
655
|
Mass spectrometric characterization of a three-enzyme tandem reaction for assembly and modification of the ...
|
|
656
|
Mass spectrometric characterization of a three-enzyme tandem reaction for assembly and modification of the ...
|
|
657
|
Mass spectrometric characterization of a three-enzyme tandem reaction for assembly and modification of the ...
|
|
658
|
Mass spectrometric characterization of a three-enzyme tandem reaction for assembly and modification of the ...
|
|
659
|
Mass spectrometric characterization of a three-enzyme tandem reaction for assembly and modification of the ...
|
|
660
|
A new kinetic model of recombinant beta-galactosidase from Kluyveromyces lactis for both hydrolysis and ...
|
|
661
|
A new kinetic model of recombinant beta-galactosidase from Kluyveromyces lactis for both hydrolysis and ...
|
|
662
|
A new kinetic model of recombinant beta-galactosidase from Kluyveromyces lactis for both hydrolysis and ...
|
|
663
|
The Effect of Adenylic Acid on Yeast Nicotinamide Adenine Dinucleotide Isocitrate Dehydrogenase, a Possible ...
|
|
664
|
Kinetic Mechanism of Histone Acetyltransferase GCN5 from Yeast
|
|
665
|
Kinetic Mechanism of Histone Acetyltransferase GCN5 from Yeast
|
|
666
|
Kinetic Mechanism of Histone Acetyltransferase GCN5 from Yeast
|
|
667
|
Kinetic Mechanism of Histone Acetyltransferase GCN5 from Yeast
|
|
668
|
Kinetic Mechanism of Histone Acetyltransferase GCN5 from Yeast
|
|
669
|
Kinetic Mechanism of Histone Acetyltransferase GCN5 from Yeast
|
|
670
|
Kinetic studies on the mechanism and regulation of rabbit liver fructose-1,6 -bisphosphatase
|
|
671
|
Kinetic studies on the mechanism and regulation of rabbit liver fructose-1,6 -bisphosphatase
|
|
672
|
Kinetic studies on the mechanism and regulation of rabbit liver fructose-1,6 -bisphosphatase
|
|
673
|
Kinetic studies on the mechanism and regulation of rabbit liver fructose-1,6 -bisphosphatase
|
|
674
|
Human brain 6-phosphogluconate dehydrogenase: purification and kinetic properties
|
|
675
|
Human brain 6-phosphogluconate dehydrogenase: purification and kinetic properties
|
|
676
|
Human brain 6-phosphogluconate dehydrogenase: purification and kinetic properties
|
|
677
|
Human brain 6-phosphogluconate dehydrogenase: purification and kinetic properties
|
|
678
|
Human brain 6-phosphogluconate dehydrogenase: purification and kinetic properties
|
|
679
|
Human brain 6-phosphogluconate dehydrogenase: purification and kinetic properties
|
|
680
|
Human brain 6-phosphogluconate dehydrogenase: purification and kinetic properties
|
|
681
|
Human brain 6-phosphogluconate dehydrogenase: purification and kinetic properties
|
|
682
|
Human brain 6-phosphogluconate dehydrogenase: purification and kinetic properties
|
|
683
|
Human brain 6-phosphogluconate dehydrogenase: purification and kinetic properties
|
|
684
|
Human brain 6-phosphogluconate dehydrogenase: purification and kinetic properties
|
|
685
|
Human brain 6-phosphogluconate dehydrogenase: purification and kinetic properties
|
|
686
|
Novel type of glucose-6-phosphate isomerase in the hyperthermophilic archaeon Pyrococcus furiosus
|
|
687
|
Novel type of glucose-6-phosphate isomerase in the hyperthermophilic archaeon Pyrococcus furiosus
|
|
688
|
Novel type of glucose-6-phosphate isomerase in the hyperthermophilic archaeon Pyrococcus furiosus
|
|
689
|
Novel type of glucose-6-phosphate isomerase in the hyperthermophilic archaeon Pyrococcus furiosus
|
|
690
|
Identifcation and functional characterization of leukotriene B4 20- hydroxylase of human polymorphonuclear ...
|
|
691
|
Identifcation and functional characterization of leukotriene B4 20- hydroxylase of human polymorphonuclear ...
|
|
696
|
Purification and characterization of avian dopamine beta-hydroxylase
|
|
697
|
Purification and characterization of avian dopamine beta-hydroxylase
|
|
698
|
Purification and characterization of avian dopamine beta-hydroxylase
|
|
699
|
Multiple forms of Human Red Blood Cell Hexokinase
|
|
700
|
Multiple forms of Human Red Blood Cell Hexokinase
|
|
701
|
Multiple forms of Human Red Blood Cell Hexokinase
|
|
702
|
Multiple forms of Human Red Blood Cell Hexokinase
|
|
703
|
Multiple forms of Human Red Blood Cell Hexokinase
|
|
704
|
Multiple forms of Human Red Blood Cell Hexokinase
|
|
705
|
Multiple forms of Human Red Blood Cell Hexokinase
|
|
706
|
Multiple forms of Human Red Blood Cell Hexokinase
|
|
707
|
Multiple forms of Human Red Blood Cell Hexokinase
|
|
708
|
Multiple forms of Human Red Blood Cell Hexokinase
|
|
709
|
Multiple forms of Human Red Blood Cell Hexokinase
|
|
710
|
Multiple forms of Human Red Blood Cell Hexokinase
|
|
711
|
Multiple forms of Human Red Blood Cell Hexokinase
|
|
712
|
Multiple forms of Human Red Blood Cell Hexokinase
|
|
713
|
Multiple forms of Human Red Blood Cell Hexokinase
|
|
714
|
Multiple forms of Human Red Blood Cell Hexokinase
|
|
715
|
Multiple forms of Human Red Blood Cell Hexokinase
|
|
716
|
Multiple forms of Human Red Blood Cell Hexokinase
|
|
717
|
Multiple forms of Human Red Blood Cell Hexokinase
|
|
718
|
Multiple forms of Human Red Blood Cell Hexokinase
|
|
719
|
Multiple forms of Human Red Blood Cell Hexokinase
|
|
720
|
NAD-dependent Methylenetetrahydrofolate Dehydrogenase-Methenyltetrahydrofolate Cyclohydrolase from Ascites ...
|
|
721
|
Probing the location and function of the conserved histidine residue of phosphoglucose isomerase by using an ...
|
|
722
|
Probing the location and function of the conserved histidine residue of phosphoglucose isomerase by using an ...
|
|
723
|
Probing the location and function of the conserved histidine residue of phosphoglucose isomerase by using an ...
|
|
724
|
Probing the location and function of the conserved histidine residue of phosphoglucose isomerase by using an ...
|
|
725
|
Probing the location and function of the conserved histidine residue of phosphoglucose isomerase by using an ...
|
|
726
|
Probing the location and function of the conserved histidine residue of phosphoglucose isomerase by using an ...
|
|
727
|
Evidence for the involvement of arginyl residue at the active site of penicillin G acylase from Kluyvera ...
|
|
728
|
Evidence for the involvement of arginyl residue at the active site of penicillin G acylase from Kluyvera ...
|
|
729
|
Evidence for the involvement of arginyl residue at the active site of penicillin G acylase from Kluyvera ...
|
|
730
|
Permeabilization of animal cells for kinetic studies of intracellular enzymes: In situ behavior of the ...
|
|
731
|
Permeabilization of animal cells for kinetic studies of intracellular enzymes: In situ behavior of the ...
|
|
732
|
Permeabilization of animal cells for kinetic studies of intracellular enzymes: In situ behavior of the ...
|
|
733
|
Permeabilization of animal cells for kinetic studies of intracellular enzymes: In situ behavior of the ...
|
|
734
|
Permeabilization of animal cells for kinetic studies of intracellular enzymes: In situ behavior of the ...
|
|
735
|
Permeabilization of animal cells for kinetic studies of intracellular enzymes: In situ behavior of the ...
|
|
736
|
Permeabilization of animal cells for kinetic studies of intracellular enzymes: In situ behavior of the ...
|
|
737
|
Permeabilization of animal cells for kinetic studies of intracellular enzymes: In situ behavior of the ...
|
|
738
|
Permeabilization of animal cells for kinetic studies of intracellular enzymes: In situ behavior of the ...
|
|
739
|
Permeabilization of animal cells for kinetic studies of intracellular enzymes: In situ behavior of the ...
|
|
740
|
Permeabilization of animal cells for kinetic studies of intracellular enzymes: In situ behavior of the ...
|
|
741
|
Permeabilization of animal cells for kinetic studies of intracellular enzymes: In situ behavior of the ...
|
|
742
|
Permeabilization of animal cells for kinetic studies of intracellular enzymes: In situ behavior of the ...
|
|
743
|
Permeabilization of animal cells for kinetic studies of intracellular enzymes: In situ behavior of the ...
|
|
744
|
Permeabilization of animal cells for kinetic studies of intracellular enzymes: In situ behavior of the ...
|
|
745
|
Permeabilization of animal cells for kinetic studies of intracellular enzymes: In situ behavior of the ...
|
|
746
|
Permeabilization of animal cells for kinetic studies of intracellular enzymes: In situ behavior of the ...
|
|
747
|
Permeabilization of animal cells for kinetic studies of intracellular enzymes: In situ behavior of the ...
|
|
748
|
Permeabilization of animal cells for kinetic studies of intracellular enzymes: In situ behavior of the ...
|
|
749
|
Permeabilization of animal cells for kinetic studies of intracellular enzymes: In situ behavior of the ...
|
|
750
|
Permeabilization of animal cells for kinetic studies of intracellular enzymes: In situ behavior of the ...
|
|
751
|
Permeabilization of animal cells for kinetic studies of intracellular enzymes: In situ behavior of the ...
|
|
752
|
Permeabilization of animal cells for kinetic studies of intracellular enzymes: In situ behavior of the ...
|
|
753
|
Identification and mechanism of a bacterial hydrolyzing UDP-N-acetylglucosamine 2-epimerase.
|
|
760
|
Purification and Properties of Human Erythrocyte Membrane NADH- Cytochrome b5 Reductase
|
|
761
|
Purification and Properties of Human Erythrocyte Membrane NADH- Cytochrome b5 Reductase
|
|
762
|
Purification and Properties of Human Erythrocyte Membrane NADH- Cytochrome b5 Reductase
|
|
763
|
Purification and Properties of Human Erythrocyte Membrane NADH- Cytochrome b5 Reductase
|
|
782
|
Structure-based engineering of Alcaligenes xylosoxidans copper-containing nitrite reductase enhances ...
|
|
783
|
Structure-based engineering of Alcaligenes xylosoxidans copper-containing nitrite reductase enhances ...
|
|
784
|
Structure-based engineering of Alcaligenes xylosoxidans copper-containing nitrite reductase enhances ...
|
|
785
|
Structure-based engineering of Alcaligenes xylosoxidans copper-containing nitrite reductase enhances ...
|
|
786
|
Structure-based engineering of Alcaligenes xylosoxidans copper-containing nitrite reductase enhances ...
|
|
787
|
Structure-based engineering of Alcaligenes xylosoxidans copper-containing nitrite reductase enhances ...
|
|
788
|
Structure-based engineering of Alcaligenes xylosoxidans copper-containing nitrite reductase enhances ...
|
|
789
|
Structure-based engineering of Alcaligenes xylosoxidans copper-containing nitrite reductase enhances ...
|
|
790
|
Structure-based engineering of Alcaligenes xylosoxidans copper-containing nitrite reductase enhances ...
|
|
817
|
Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures ...
|
|
818
|
Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures ...
|
|
819
|
Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures ...
|
|
820
|
Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures ...
|
|
821
|
Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures ...
|
|
822
|
Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures ...
|
|
823
|
Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures ...
|
|
824
|
Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures ...
|
|
825
|
Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures ...
|
|
826
|
Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures ...
|
|
905
|
Two novel mutant human adenylosuccinate lyases (ASLs) associated with autism and characterization of the ...
|
|
906
|
Two novel mutant human adenylosuccinate lyases (ASLs) associated with autism and characterization of the ...
|
|
907
|
Two novel mutant human adenylosuccinate lyases (ASLs) associated with autism and characterization of the ...
|
|
908
|
Two novel mutant human adenylosuccinate lyases (ASLs) associated with autism and characterization of the ...
|
|
909
|
Two novel mutant human adenylosuccinate lyases (ASLs) associated with autism and characterization of the ...
|
|
910
|
Two novel mutant human adenylosuccinate lyases (ASLs) associated with autism and characterization of the ...
|
|
911
|
Two novel mutant human adenylosuccinate lyases (ASLs) associated with autism and characterization of the ...
|
|
912
|
Two novel mutant human adenylosuccinate lyases (ASLs) associated with autism and characterization of the ...
|
|
913
|
Two novel mutant human adenylosuccinate lyases (ASLs) associated with autism and characterization of the ...
|
|
914
|
Two novel mutant human adenylosuccinate lyases (ASLs) associated with autism and characterization of the ...
|
|
915
|
Two novel mutant human adenylosuccinate lyases (ASLs) associated with autism and characterization of the ...
|
|
916
|
Two novel mutant human adenylosuccinate lyases (ASLs) associated with autism and characterization of the ...
|
|
917
|
Two novel mutant human adenylosuccinate lyases (ASLs) associated with autism and characterization of the ...
|
|
918
|
Two novel mutant human adenylosuccinate lyases (ASLs) associated with autism and characterization of the ...
|
|
919
|
Two novel mutant human adenylosuccinate lyases (ASLs) associated with autism and characterization of the ...
|
|
920
|
Two novel mutant human adenylosuccinate lyases (ASLs) associated with autism and characterization of the ...
|
|
921
|
Two novel mutant human adenylosuccinate lyases (ASLs) associated with autism and characterization of the ...
|
|
922
|
Two novel mutant human adenylosuccinate lyases (ASLs) associated with autism and characterization of the ...
|
|
923
|
Two novel mutant human adenylosuccinate lyases (ASLs) associated with autism and characterization of the ...
|
|
924
|
Two novel mutant human adenylosuccinate lyases (ASLs) associated with autism and characterization of the ...
|
|
925
|
Two novel mutant human adenylosuccinate lyases (ASLs) associated with autism and characterization of the ...
|
|
926
|
Two novel mutant human adenylosuccinate lyases (ASLs) associated with autism and characterization of the ...
|
|
927
|
Two novel mutant human adenylosuccinate lyases (ASLs) associated with autism and characterization of the ...
|
|
928
|
Two novel mutant human adenylosuccinate lyases (ASLs) associated with autism and characterization of the ...
|
|
933
|
Active site mutants of the "non-hydrolyzing" UDP-N-acetylglucosamine 2-epimerase from Escherichia coli
|
|
934
|
Active site mutants of the "non-hydrolyzing" UDP-N-acetylglucosamine 2-epimerase from Escherichia coli
|
|
935
|
Active site mutants of the "non-hydrolyzing" UDP-N-acetylglucosamine 2-epimerase from Escherichia coli
|
|
936
|
Active site mutants of the "non-hydrolyzing" UDP-N-acetylglucosamine 2-epimerase from Escherichia coli
|
|
937
|
Active site mutants of the "non-hydrolyzing" UDP-N-acetylglucosamine 2-epimerase from Escherichia coli
|
|
938
|
Active site mutants of the "non-hydrolyzing" UDP-N-acetylglucosamine 2-epimerase from Escherichia coli
|
|
939
|
Active site mutants of the "non-hydrolyzing" UDP-N-acetylglucosamine 2-epimerase from Escherichia coli
|
|
940
|
Active site mutants of the "non-hydrolyzing" UDP-N-acetylglucosamine 2-epimerase from Escherichia coli
|
|
941
|
Active site mutants of the "non-hydrolyzing" UDP-N-acetylglucosamine 2-epimerase from Escherichia coli
|
|
942
|
Active site mutants of the "non-hydrolyzing" UDP-N-acetylglucosamine 2-epimerase from Escherichia coli
|
|
943
|
Catalytic mechanism of alpha-retaining glucosyl transfer by Corynebacterium callunae starch phosphorylase: the ...
|
|
944
|
Catalytic mechanism of alpha-retaining glucosyl transfer by Corynebacterium callunae starch phosphorylase: the ...
|
|
945
|
Catalytic mechanism of alpha-retaining glucosyl transfer by Corynebacterium callunae starch phosphorylase: the ...
|
|
946
|
Catalytic mechanism of alpha-retaining glucosyl transfer by Corynebacterium callunae starch phosphorylase: the ...
|
|
947
|
Catalytic mechanism of alpha-retaining glucosyl transfer by Corynebacterium callunae starch phosphorylase: the ...
|
|
948
|
Catalytic mechanism of alpha-retaining glucosyl transfer by Corynebacterium callunae starch phosphorylase: the ...
|
|
949
|
Catalytic mechanism of alpha-retaining glucosyl transfer by Corynebacterium callunae starch phosphorylase: the ...
|
|
950
|
Catalytic mechanism of alpha-retaining glucosyl transfer by Corynebacterium callunae starch phosphorylase: the ...
|
|
951
|
Catalytic mechanism of alpha-retaining glucosyl transfer by Corynebacterium callunae starch phosphorylase: the ...
|
|
952
|
Catalytic mechanism of alpha-retaining glucosyl transfer by Corynebacterium callunae starch phosphorylase: the ...
|
|
953
|
Catalytic mechanism of alpha-retaining glucosyl transfer by Corynebacterium callunae starch phosphorylase: the ...
|
|
954
|
Catalytic mechanism of alpha-retaining glucosyl transfer by Corynebacterium callunae starch phosphorylase: the ...
|
|
955
|
Catalytic mechanism of alpha-retaining glucosyl transfer by Corynebacterium callunae starch phosphorylase: the ...
|
|
956
|
Catalytic mechanism of alpha-retaining glucosyl transfer by Corynebacterium callunae starch phosphorylase: the ...
|
|
957
|
Catalytic mechanism of alpha-retaining glucosyl transfer by Corynebacterium callunae starch phosphorylase: the ...
|
|
958
|
Catalytic mechanism of alpha-retaining glucosyl transfer by Corynebacterium callunae starch phosphorylase: the ...
|
|
959
|
Catalytic mechanism of alpha-retaining glucosyl transfer by Corynebacterium callunae starch phosphorylase: the ...
|
|
960
|
Catalytic mechanism of alpha-retaining glucosyl transfer by Corynebacterium callunae starch phosphorylase: the ...
|
|
961
|
Catalytic mechanism of alpha-retaining glucosyl transfer by Corynebacterium callunae starch phosphorylase: the ...
|
|
962
|
Catalytic mechanism of alpha-retaining glucosyl transfer by Corynebacterium callunae starch phosphorylase: the ...
|
|
963
|
Catalytic mechanism of alpha-retaining glucosyl transfer by Corynebacterium callunae starch phosphorylase: the ...
|
|
964
|
Catalytic mechanism of alpha-retaining glucosyl transfer by Corynebacterium callunae starch phosphorylase: the ...
|
|
965
|
Catalytic mechanism of alpha-retaining glucosyl transfer by Corynebacterium callunae starch phosphorylase: the ...
|
|
966
|
The carboligation reaction of acetohydroxyacid synthase II: steady-state intermediate distributions in wild ...
|
|
967
|
The carboligation reaction of acetohydroxyacid synthase II: steady-state intermediate distributions in wild ...
|
|
968
|
The carboligation reaction of acetohydroxyacid synthase II: steady-state intermediate distributions in wild ...
|
|
969
|
The carboligation reaction of acetohydroxyacid synthase II: steady-state intermediate distributions in wild ...
|
|
970
|
The carboligation reaction of acetohydroxyacid synthase II: steady-state intermediate distributions in wild ...
|
|
971
|
Inhibition of type I and type II phosphomannose isomerases by the reaction intermediate analogue ...
|
|
972
|
Inhibition of type I and type II phosphomannose isomerases by the reaction intermediate analogue ...
|
|
973
|
Inhibition of type I and type II phosphomannose isomerases by the reaction intermediate analogue ...
|
|
974
|
Inhibition of type I and type II phosphomannose isomerases by the reaction intermediate analogue ...
|
|
979
|
Identification, characterization, and site-directed mutagenesis of recombinant pentachlorophenol ...
|
|
980
|
Identification, characterization, and site-directed mutagenesis of recombinant pentachlorophenol ...
|
|
981
|
Identification, characterization, and site-directed mutagenesis of recombinant pentachlorophenol ...
|
|
982
|
Identification, characterization, and site-directed mutagenesis of recombinant pentachlorophenol ...
|
|
983
|
Identification, characterization, and site-directed mutagenesis of recombinant pentachlorophenol ...
|
|
984
|
Identification, characterization, and site-directed mutagenesis of recombinant pentachlorophenol ...
|
|
985
|
Nikkomycin biosynthesis: formation of a 4-electron oxidation product during turnover of NikD with its ...
|
|
986
|
Nikkomycin biosynthesis: formation of a 4-electron oxidation product during turnover of NikD with its ...
|
|
987
|
The Catalytic Mechanism of the Glutathione-Dependent Dehydroascorbate Reductase Activity of Thioltransferase ...
|
|
988
|
The Catalytic Mechanism of the Glutathione-Dependent Dehydroascorbate Reductase Activity of Thioltransferase ...
|
|
989
|
The Catalytic Mechanism of the Glutathione-Dependent Dehydroascorbate Reductase Activity of Thioltransferase ...
|
|
990
|
The Catalytic Mechanism of the Glutathione-Dependent Dehydroascorbate Reductase Activity of Thioltransferase ...
|
|
991
|
Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the ...
|
|
992
|
Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the ...
|
|
993
|
Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the ...
|
|
994
|
Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the ...
|
|
995
|
Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the ...
|
|
996
|
Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the ...
|
|
997
|
Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the ...
|
|
998
|
Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the ...
|
|
999
|
Structural studies of the catalytic reaction pathway of a hyperthermophilic histidinol-phosphate ...
|
|
1000
|
Structural studies of the catalytic reaction pathway of a hyperthermophilic histidinol-phosphate ...
|





 If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.
If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.